From infection to ARDS: the role of the gut microbiome in SARS-COV2
A schematic, limited hypothesis outlining how gut dysbiosis from a SARS-COV2 infection may proceed to sepsis and ARDS.
The following post will try to piece together the available information to formalize a hypothesis on the role of a SARS-COV2 infection on gut dysbiosis, gut translocation, and possibly the exacerbation of sepsis and ARDS. Note that the following is a simplified hypothesis with many missing pieces, but collects what was previously discussed.
Edit 1/16/2023: An additional Figure and citation (from Mukherjee, S., & Hanidziar, D., Footnote 11) was included for point 3 below, which details the points made very well by explaining how gut microbes may translocate to the lungs and provide further damage and the culmination into ARDS.
In conclusion to Bernard-Raichon, et al.
The study from Bernard-Raichon, et al.1 (discussed in the previous article) appeared, at first, to be a possible smoking gun in tying together the secondary bacterial infections seen in hospitalized SARS-COV2 patients.
However, upon closer inspection it missed out on a lot of necessary details. Unfortunately, the remarks about nosocomial infections were a bit of a red herring, as the actual body of work didn’t provide substantial evidence, just observations that gut bacteria were found in a few patients, but not how they got there or what this would actually mean as the bacteremia with gut bacteria were never associated with any respective septic or deceased patient.
Of course, that’s how scientific studies go sometimes.
One can consider a study as a snapshot into the play that is SARS-COV2. All the players are on stage, and yet you catch only a mere glimpse of the event as it occurs in one experiment. Another experiment may provide another snapshot where the actors are rearranged, and yet you may not know what actions led to them being where they are.
Essentially, studies are either still images or just brief glimpses into the process at work, stitched together through different experiments and methodologies in order to try to recreate the play.
In any given case, there will always be gaps in the stitching together of information. In some cases you may realize that the actors aren’t exactly where they should be (or where you assume they should be) or that one person’s picture provides a different perspective than your own.
There’s a reason why the scientific process can be analogized to a crime scene, drawing together enough evidence to find someone (or something) culpable for the event. But rather than using a “beyond a reasonable doubt standard”, science tends to relegate itself towards the “preponderance of evidence”. That is, “50% + X” is enough, even if “X” here is a mere 0.00001%.
And so, in piecing together the information understand that we are only provided snapshots into the events as they unfold, having to fill in the gaps with our own biases or assumptions to act as the tying feature between each experiment.
A hypothetical model for the gut microbiome’s role in SARS-COV2 and sepsis/ARDS
With that, know that the following hypothetical is constructed based on the “images” taken from various studies with serious gaps in information. It most likely doesn’t even fit the “preponderance of evidence”, and only serves as one multitude of possibilities out there. However, the construction of this hypothesis is more done to provide a possibility, and show a process outside of the presentation of still images alone.
So we will trace a select pathway, detailing how an infection with SARS-COV2 will progress to gut dysfunction and microbiome alterations, leading to passage of gut bacteria into the blood, which will then make their way to the damaged lungs and cause secondary bacterial infections.
1. A SARS-COV2 infection takes hold
Let’s hypothesize a random individual who has come into contact with a SARS-COV2 infected individual and themselves have become infected. The person will develop progressively worse symptoms resulting in eventual ARDS and death.
At the early onset of the infection the virus makes its way down the respiratory tract into the lungs of the infected individual.
There, the virus targets the microenvironment, infecting the host cells by binding to ACEII receptors on the surface of alveolar epithelial cells and damaging the local tissue.
At the same point that this infection begins to spread and take hold, the body mounts an immune response with a primary innate immune response followed by humoral and cellular immune responses, all of which result in the release of inflammatory molecules such as cytokines and chemokines intended to activate the necessary immune cells.2
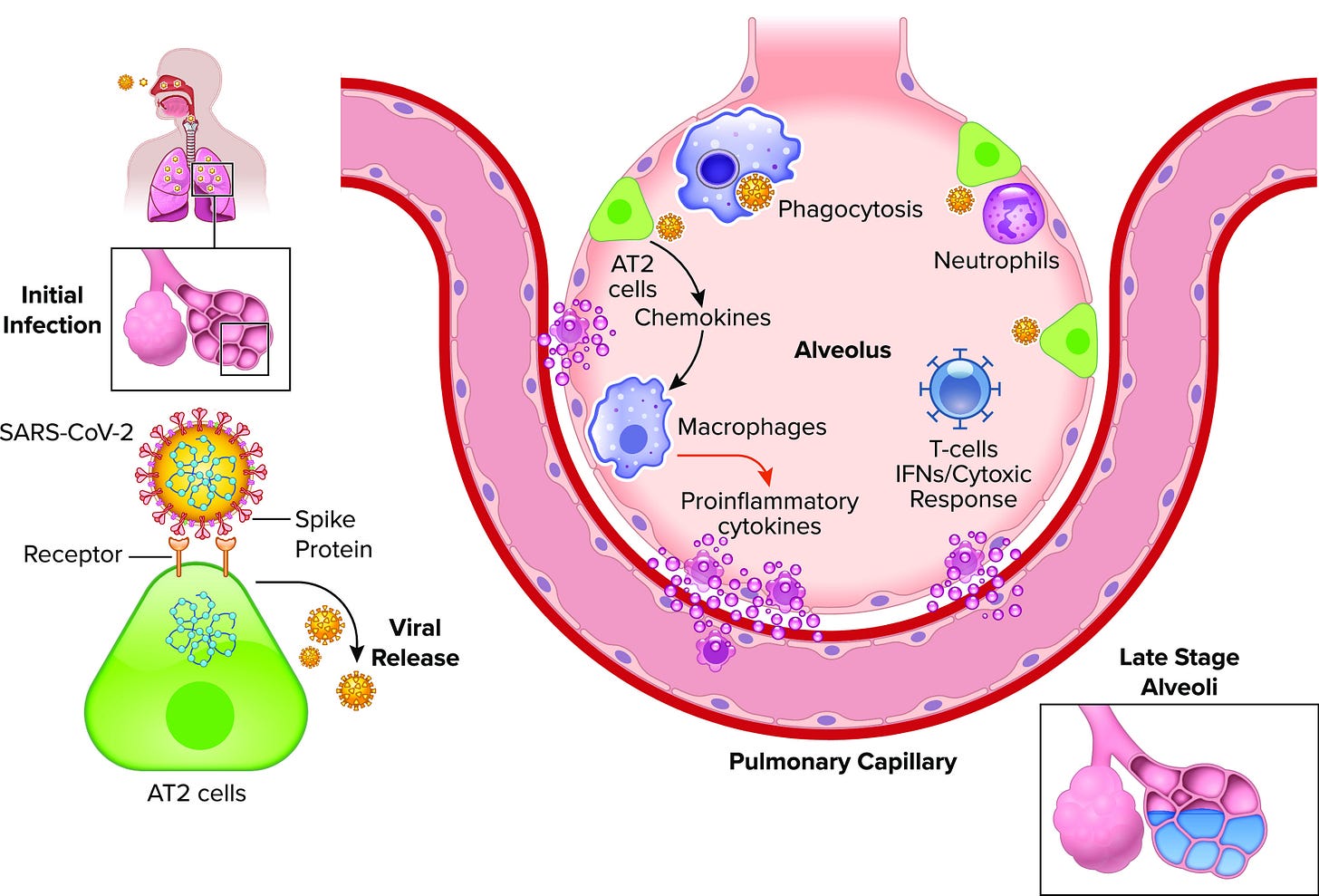
Unfortunately, at the point that inflammatory molecules are released the body must now engage in a balancing act. Although the immune system is now activated to deal with the virus, the inflammatory molecules themselves kill off cells, inflame the lung tissues, and alter the lung microbiome.3
For instance, as monocytes and macrophages are recruited into the local environment they release inflammatory cytokines, which may then recruit neutrophils. As they degranulate they may further damage the endothelial cells of the lungs as they attempt to target the virus.4
Concurrently, as various immune cells continue to make their way from the lymphatic system into the bloodstream and then into infected alveoli, the cells loosen the tight junctions and become more permeable. Now, content within the alveoli may enter into the bloodstream, including cytokines and other inflammatory markers. As these inflammatory markers circulate they begin to induce inflammatory responses in distant regions of the body, and therefore the inflammation moves from localized to systemic.
2. Inflammation and possibly direct viral infection alter the gut microbiome
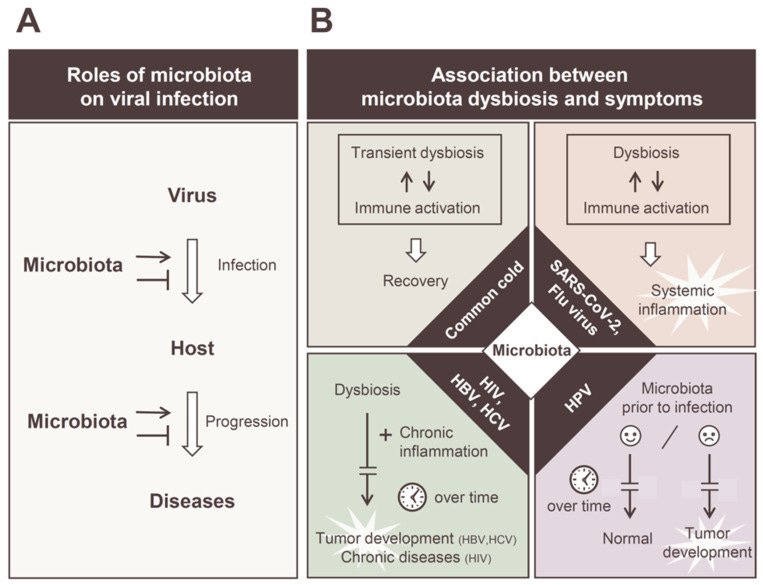
*Footnote for Mizutani, et al.5
As inflammatory markers such as TNF-alpha, interleukins, and antiviral molecules such as interferons circulate within the bloodstream they may make their way to the gut where such inflammatory markers induce alterations in the gut microbes.6
This may also occur as the virus possibly infects the epithelial cells of the gut as well, producing similar effects as those seen within the lungs.
Microbes susceptible to these inflammatory molecules may decrease in numbers while others increase. This also comes as the body may suffer from a reduction in oxygen due to lung injury, as well as inappetence (lack of appetite) that alters the available nutrients for the gut microbes such as dietary fiber.
As such, the gut environment begins to deteriorate, with beneficial bacteria such as Faecalibacterium on the decline while other bacteria such as Akkermansia, Bacteroides, and E. coli may increase.
At this point, the alterations to the gut may erode the mucosal barrier, the gut becomes inflamed, and permeability results here as well.7 The loss of beneficial bacterial, as well as inappetence, may reduce the production of anti-inflammatory short chain fatty acids responsible for attenuating the inflammatory response coming from the lungs and now the gastrointestinal tract.
Now, the gut may have to deal with its own dysbiosis by mounting an immune response to deal with the increase in pathogenic bacteria as well as the possible viral infection, and thus a feedback loop may be initiated that continues to promote systemic inflammation.8
If uncontrolled, the damage will become severe.

As the gut is now made more permeable, opportunistic bacteria may translocate from the gut into the blood stream and begin circulating towards other organs such as the liver, heart, or even the lungs.
Now that bacteria may enter the blood, the individual may now begin to experience symptoms of sepsis.9
Of course, if the infection is too severe at this point, the individual may also become hospitalized, in which case antibiotics may be prescribed that further exacerbate the dysbiosis, while also possibly selecting for antibiotic-resistant bacteria in the process.
Factor in the likelihood that the individual may be administered intravenous solutions, and possibly intubated and the risk of nosocomial infections increases, with several of these bacteria finding access into the bloodstream as noticed with intravenous catheters.10
3. Sepsis develops as bacteria colonize damaged lungs
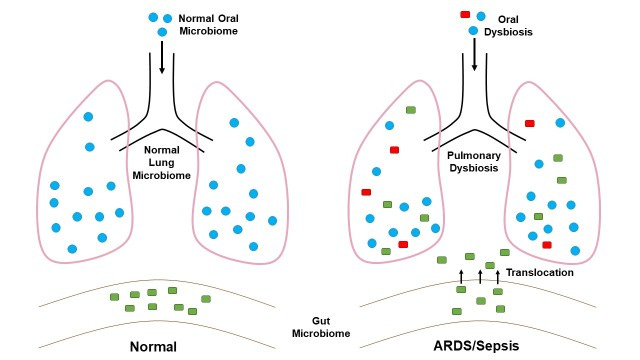
Now that the patient is experiencing bacteremia the bacteria may spread to other regions of the body. The lungs, being damaged by SARS-COV2 and the excessive inflammation, are susceptible to colonization by bacteria.11
Thus, bacteria in the blood may translocate into the lungs, where the lower oxygen levels may now be more hospitable to select anaerobic species.
Afterwards, a secondary bacterial infection may arise. As the body now has to deal with both a viral infection as well as a bacterial infection, the body’s defenses may be overwhelmed with attempting to quell the continuous bombardments.
The end result may be bacterial pneumonia or exacerbation of lung damage that may add to the possibility of ARDS, and the eventual death of the patient.12
Major caveats to the hypothesis
Of course, the hypothesis is just that: a mere hypothesis collecting bits of information, taking images and attempting to construct a narrative in order to provide a conjecture. It’s also important to point out that this hypothesis is not original, given that it derives from studies that also support this idea.
The evidence here is lacking in many places, and is a more simplified summary than was originally planned.
Note that the strains of bacteria mentioned here may not be the actual players in ARDS or sepsis related to a SARS-COV2 infection. The ones cited were based on some of the articles mentioned in previous Substack posts that detail these bacteria in particular. However, it’s more than likely that the key bacteria may be of different species. In particular, bacterial pneumonia stems mostly from, of course, Streptococcus pneumonia13, and so that raises the question as to whether this same species should be found in secondary bacterial infections with SARS-COV2 hospitalized patients.
We also don’t have much evidence as to how the bacteria will travel from the gut to the lungs. Here, I posited that the circulatory system may be a possible route given the evidence of bacteremia from the Bernard-Raichon, et al. study, although the researchers there also provide a conjecture by inferring gut permeability but without providing significant evidence (again, in my opinion). This appears to be a good possibility given that bacteremia is well-noted within the literature in those with viral infections, as well as other diseases. Gut bacteria may just travel the route of the digestive tract as well to make their way, but again that is more conjecture without any evidence.
Also, not all cases of severe COVID result in secondary bacterial infection. It’s quite possible that the extreme inflammatory response, possibly owed in part to gut dysbiosis without necessary gut translocation, may suffice in the cytokine storms seen.
SARS-COV2 can cause enough damage to the lungs itself without requiring additional pathologies.
There’s more to discuss that’s worth considering, such as the role of the hospital in contributing to secondary bacterial infections.
But that will be saved for another time. Again, remember that the above is a mere hypothesis that may relate to a select few individuals.
If you enjoyed this post and other works please consider supporting me through a paid Substack subscription or through my Ko-fi. Any bit helps, and it encourages independent creators and journalists outside the mainstream.

Bernard-Raichon, L., Venzon, M., Klein, J., Axelrad, J. E., Zhang, C., Sullivan, A. P., Hussey, G. A., Casanovas-Massana, A., Noval, M. G., Valero-Jimenez, A. M., Gago, J., Putzel, G., Pironti, A., Wilder, E., Yale IMPACT Research Team, Thorpe, L. E., Littman, D. R., Dittmann, M., Stapleford, K. A., Shopsin, B., … Schluter, J. (2022). Gut microbiome dysbiosis in antibiotic-treated COVID-19 patients is associated with microbial translocation and bacteremia. Nature communications, 13(1), 5926. https://doi.org/10.1038/s41467-022-33395-6
Upadhya, S., Rehman, J., Malik, A. B., & Chen, S. (2022). Mechanisms of Lung Injury Induced by SARS-CoV-2 Infection. Physiology (Bethesda, Md.), 37(2), 88–100. https://doi.org/10.1152/physiol.00033.2021
Dumas, A., Bernard, L., Poquet, Y., Lugo-Villarino, G., & Neyrolles, O. (2018). The role of the lung microbiota and the gut-lung axis in respiratory infectious diseases. Cellular microbiology, 20(12), e12966. https://doi.org/10.1111/cmi.12966
Batah, S. S., & Fabro, A. T. (2021). Pulmonary pathology of ARDS in COVID-19: A pathological review for clinicians. Respiratory medicine, 176, 106239. https://doi.org/10.1016/j.rmed.2020.106239
Mizutani, T., Ishizaka, A., Koga, M., Tsutsumi, T., & Yotsuyanagi, H. (2022). Role of Microbiota in Viral Infections and Pathological Progression. Viruses, 14(5), 950. https://doi.org/10.3390/v14050950
Sencio, V., Machado, M. G., & Trottein, F. (2021). The lung-gut axis during viral respiratory infections: the impact of gut dysbiosis on secondary disease outcomes. Mucosal immunology, 14(2), 296–304. https://doi.org/10.1038/s41385-020-00361-8
Giron, L. B., Dweep, H., Yin, X., Wang, H., Damra, M., Goldman, A. R., Gorman, N., Palmer, C. S., Tang, H. Y., Shaikh, M. W., Forsyth, C. B., Balk, R. A., Zilberstein, N. F., Liu, Q., Kossenkov, A., Keshavarzian, A., Landay, A., & Abdel-Mohsen, M. (2021). Plasma Markers of Disrupted Gut Permeability in Severe COVID-19 Patients. Frontiers in immunology, 12, 686240. https://doi.org/10.3389/fimmu.2021.686240
Zhang, F., Lau, R.I., Liu, Q. et al. Gut microbiota in COVID-19: key microbial changes, potential mechanisms and clinical applications. Nat Rev Gastroenterol Hepatol (2022). https://doi.org/10.1038/s41575-022-00698-4
Adelman, M. W., Woodworth, M. H., Langelier, C., Busch, L. M., Kempker, J. A., Kraft, C. S., & Martin, G. S. (2020). The gut microbiome's role in the development, maintenance, and outcomes of sepsis. Critical care (London, England), 24(1), 278. https://doi.org/10.1186/s13054-020-02989-1
Gahlot, R., Nigam, C., Kumar, V., Yadav, G., & Anupurba, S. (2014). Catheter-related bloodstream infections. International journal of critical illness and injury science, 4(2), 162–167. https://doi.org/10.4103/2229-5151.134184
Mukherjee, S., & Hanidziar, D. (2018). More of the Gut in the Lung: How Two Microbiomes Meet in ARDS. The Yale journal of biology and medicine, 91(2), 143–149.
*Citation added retroactively on 1/16/2023
Manohar, P., Loh, B., Nachimuthu, R., Hua, X., Welburn, S. C., & Leptihn, S. (2020). Secondary Bacterial Infections in Patients With Viral Pneumonia. Frontiers in medicine, 7, 420. https://doi.org/10.3389/fmed.2020.00420
Eshwara, V. K., Mukhopadhyay, C., & Rello, J. (2020). Community-acquired bacterial pneumonia in adults: An update. The Indian journal of medical research, 151(4), 287–302. https://doi.org/10.4103/ijmr.IJMR_1678_19



Excellent commentary, sir. It's clear you have a good grasp on this topic. Thank you for sharing.
I think the idea of widespread viral infection is a bit of a myth. I believe there is some kind of poisoning occurring that will often lead to secondary bacterial infections.