What proteins are mRNA vaccines producing?
A quick, unthorough look at the recent Nature article on frameshifted proteins due to N-methylpseudouridine.
Several people have reported on this study already (links below). I haven’t looked at it in full yet so this is a brief summary of what I have looked at so far, so please keep that in mind that the information below will be based on limited assessment.
Some people who have covered this article so far:
As several people have reported on by now, a recent study published in Nature1 appears to point to another flaw in the mRNA vaccines. In this case, it appears that some of the proteins being produced may not be analogous to the spike protein, but rather may be peptides formed by what’s called a ribosomal frameshift.
Before we begin
Frameshifts are somewhat common in both replication and translation. Most people know of frameshift mutations, in which bases may be inserted or deleted from either the reference genome or during transcription, all of which cause the downstream sequence of codons to shift over. Remember that translation, the process of forming proteins, is done in bases of 3. When the proper codon is recognized and the proper amino acid is inserted into the growing peptide strand this is called an in-frame insertion. If shifted over, this is referred to as being out of frame. Frameshift mutations can be rather disastrous when the insertion/deletion occurs in numbers not divisible by 3 because it leads to downstream codon shifts, which then lead to insertions of wrong amino acids and an overall protein that was not the initially intended product.
Translational frameshifts/ribosomal frameshifts are different than frameshift mutations, as these frameshifts are due to misreads of mRNA rather than mutations that may reside within the genetic sequence.
In this case, the end result is the same between frameshift mutations and ribosomal frameshifts as the downstream amino acid sequences end up being far different than what the intended sequence would be. However, frameshift mutations can be more harmful as translational errors may be minor relative to the entire pool of proteins produced.
As I’m not very creative Wikipedia has a layman’s example for ribosomal frameshifts:

Note here that the amino acid sequence doesn’t inherently “not make sense”. Rather, a better argument would be that it’s a mistranslation for what you would have wanted (i.e. it’s not quite in the language that you expected, which also may infer that the translated sentence won’t be useful). Amino acids are still incorporated, but they aren’t the ones you originally wanted and won’t have the initial function.
Along with wrong amino acid incorporations comes the possibility that ribosomes may also improperly frameshift and interpret stop codons where there normally wouldn’t be any, leading to truncated peptides. In this case, a shift over may lead to recognition of a stop codon that normally wouldn’t be there if the mRNA sequence was read properly, leading to the abrupt stop of translation.
In short, there are two forms of ribosomal frameshifts which are labeled somewhat counterintuitively. For the sake of this article note that the study in question refers to a +1 frameshift. The “+1” refers to the fact that, at some point during translation, the ribosome appears to shift forward one base, essentially skipping one base and causing a shift over to the right from what was originally intended. This is referred to as a “+1” frameshift if it is only one base slip to the right. There’s also the possibility of a -1 frameshift, in which the ribosome may move backwards one base which will also led to out of frame reading downstream of the frameshift.
Now, the reason for ribosomal frameshifts are not too clear. It appears that within the context of +1 frameshifts this may be a consequence of ribosomal stalling where the ribosome stops for a longer than expected amount of time. This may be due to the fact that tRNAs, structures which bear opposite codons to mRNA referred to as anticodons that help carry specific amino acids and extend the growing amino acid sequence, are not equally prevalent within cells. More “rare” tRNAs will have fewer probabilities of binding to its proper codon sequence, causing the ribosome to wait for this binding to occur. This may allow time for other tRNAs which can recognize out of frame but otherwise nearby codons to bind, leading to a frameshift.
Again, Wikipedia provides this example in which ribosomal stalling due to less prevalent Arginine-bearing tRNAs that bind to the codon CUU may allow for Glycine-bearing tRNAs to bind by recognizing a codon sequence that is one base to the right of the initial Arginine codon:

Keep in mind that the ribosomal stalling also serves to allow room for more thermodynamically favorable binding to occur, in which case ribosome bearing the previous codon may shift over to a more favorable spot and induce the frameshift (this appears to be the explanation for the scenario presented above).
Instances where ribosomal stalling takes place (as in the example above) are referred to as slippery sequences as they are regions where ribosomes may be more prone to slip and incur frameshifts.
This is an important thing to keep in mind when considering why the mRNA vaccines may be inducing frameshifts.
A brief look at the study
Again, keep in mind I haven’t looked at this study deeply so I’m only assessing things based on what I was able to understand.
First off, the researchers wanted to determine if ribosomal frameshifting was occurring with mRNA vaccines due to the fact that viruses are known to incur ribosomal frameshifts. In order to test their hypothesis the researchers utilized a reporter gene, in this case one that codes for the enzyme luciferase. Reporter genes are, by their name, genes that form proteins which can lead to detectable products. In this case, the use of a luciferase gene allows researchers to detect fluorescence by way of examining luciferase activity, with greater luciferase activity (measured through fluorescence by luciferin metabolism) inferring proper translated products (i.e. fewer ribosomal frameshifting).
In order to test this, researchers used one wildtype luciferase gene (WT Fluc- no, that doesn’t stand for “what the fluc”, although you may incorporate that into your own usage) in order to examine alterations in activity related to modified bases. In order to test the hypothesis of ribosomal frameshifts the researchers also utilized a modified sequences of luciferase. In this case, the modification was the insertion of a base that would code for a stop codon in the middle of the luciferase sequence.
This is a pretty interesting approach as all translations lacking any ribosomal frameshifts would lead to truncated luciferase since it would recognize the artificially created stop codon. Essentially, the researchers created a hurdle which can only be overcome if ribosomal frameshifting occurs.
Using the same sentence taken from Wikipedia, this would be similar as adding in a letter such as the B below that throws off the rest of the sentence, only in the case of luciferase the sentence/functional protein would be cut short:
Unmodified:
|Start|THE CAT AND THE MAN ARE FAT ...
|Start|123 123 123 123 123 123 123 ...Artificial insertion leading to an out-of-frame sequence relative to the reference luciferase gene:
|Start|THE CAT BAN DTH EMA NAR EFA ...
|Start|123 123 123 123 123 123 123 ...Note that this is a game of probability. Ribosomal frameshifting may occur at various points during the sequence/sentence, and so both a mix of truncated and properly translated luciferases should be expected. Rather, it’s the proportion of truncated or garbled luciferases (measured by reduced luciferase activity) relative to unmodified luciferase that’s important as it provides a measure of how often this frameshifting is occurring.
Put another way, in reference to the now garbled sentence above, the researchers are seeing if ribosomal frameshifts are occurring by measuring how often the inserted B would be skipped, which would then lead to the an in-frame reading, or otherwise the proper sentence/luciferase. Make the protein out-of-frame and see if the ribosomes can make them in-frame.
The results of this portion are noted below:
Note that (a) shows the normal luciferase gene compared to the gene which inserted a base to incur a stop codon (Fluc+1FS, with the stop codon noted with the *). The researchers used sequences that had regular, unmodified mRNA along with mRNA for luciferase that contained various base modifications. In this case the m1Ψ modification (denoting the methylated pseudouridine) is the important result here as they are the ones found within the mRNA vaccines, but note that various combinations of base modifications and pairings were used in this study.
(b) looks at luciferase activity when using the wildtype luciferase gene and different base modifications (shown on the bottom of the graph). In this case, we shouldn’t expect ribosomal frameshifts to occur outside of the normal rate (noted in the unmodified gene reference).
Interestingly, the use of wildtype luciferase modifications with m1Ψ appear to show reduced luciferase activity (argued to not be statistically significant) relative to the unmodified sequence. This would at least infer that something is happening during translation that is reducing the number of functional luciferase. There are several factors here, but the main assumption would of course be that some sort of frameshifting is occurring and causing non-luciferase proteins to be formed, or if formed are possibly truncated and non-functioning.
In (c) the gene sequence bearing the additional base (Fluc+1FS) is used, and here there’s a rather striking finding. In this case all of the sequences showed no luciferase activity aside from the sequence bearing the m1Ψ modifications. Again, remember that if no ribosomal frameshifts occurred we would expect no luciferase activity. Therefore, the fact that the sequence bearing the m1Ψ modification showed luciferase activity we can infer that some form of ribosomal frameshifting has occurred.
However, note that the scaling still suggests that not many of the ribosomes are able to overcome the hurdle. The unmodified sequence for the wildtype luciferase (Figure 1b) show activity around 15 million while the m1Ψ-bearing sequence shows activity around 10 million. In contrast, the luciferase sequence with the hurdle base shows activity somewhere around 1 million. Although there is still activity, there’s still a relative decline of 1/10 in luciferase activity, again suggesting that even if these ribosomal frameshifts are occurring it may be relatively rare. Remember, “relatively” is the important word here given that only 3 assays appear to be done across the study, as well as the fact that this portion of the study was done in vitro.
Also, note that luciferase activity was also lower when HeLa cells were transfected with the Fluc+1FS gene rather than free ribosomes, although this may be due to rates of transfection. Unfortunately, no wildtype sequence was provided as a comparison to see what type of activity would normally be expected.
Now, given these in vitro findings the authors turned to mice models to examine the extent of possible ribosomal frameshifts.
Remember that the intent of this study is to consider the possibility that mRNA vaccines may lead to +1 ribosomal frameshifting, and therefore individuals or animal models provided these vaccines should become sensitized to out-of-frame peptide products derived from the spike.
Here, mice were either left unvaccinated, provided Pfizer/BioNTech’s mRNA vaccine, or given the adenoviral-based vaccine. The use of the adenoviral-based vaccine serves to see whether this ribosomal frameshifting may be occurring in vaccines bearing unmodified bases.
It’s important to note that the mice in this study were given 3 doses of each vaccine rather than the typical 2 dose that were initially considered “fully vaccinated” in humans:
Mice of 8–12 weeks old were intramuscularly injected with three doses of 10 μg BNT162b2, 5 × 107 infectious units of ChAdOx1 nCoV-19 or untreated. For booster immunizations, the same dose of the respective vaccine was injected 3 weeks and 6 weeks apart into the same site as the primary immunization.
After vaccination mice (at least cells derived from their spleens) were challenged with peptides derived from spike and peptides which are alleged to occur from +1 ribosomal frameshifts. Predict is the important word here, as the researchers didn’t necessarily take the vaccines and induce translation and isolate possible +1 ribosomal frameshift products. It appears that they used an in silico method of finding sequences and outsourced production of these out-of-frame peptides (emphasis mine):
To address this possibility, we vaccinated mice with BNT162b2 and quantified their T cell response to in-frame SARS-CoV-2 spike protein and +1 frameshifted products predicted to occur by translation of the mRNA +1 frame, as well as an unrelated control antigen (SARS-CoV-2 M protein), by interferon-γ (IFNγ) ELISpot assay.
Note that this pool of peptides used is rather exhaustive, as noted in the following table. +1FS spike refer to those peptides predicted to come from +1 ribosomal frameshifting:
Also, keep in mind that the researchers are examining immune responses by looking at T-cells. In particular, they are measuring interferon-γ (IFNγ) responses, which may infer immune system activity.
The results of this portion are noted below:
Note that the graph on the left is showing responses to the predicted out-of-frame peptides across the different mice groups. The SARS-CoV-2 M group seems to have been thrown in as another negative control. The right graph examines these same mice’s responses to peptides derived from the in-frame predicted spike peptides i.e. responses that should be expected towards the spike.
In both vaccinated groups mice show a clear IFNγ response to in-frame spike peptides. The untreated mice appear to show limited responses, although remember to watch out for scaling (the y-axis for (c) appears logarithmic) and so the responses seen in untreated mice may be noise.
What’s important for our sake is the response to the +1 frameshift predicted peptides (Figure 2b). Here, there appears to be a clear, distinct IFNγ response in mRNA-vaccinated mice, suggesting that possible frameshifting may be occurring.
It’s important to note that, similar to the in vitro models, the difference in responses are by orders of magnitude, which would be expected as ribosomal frameshifting should be occurring abundantly. Because of the logarithmic graph for the spike it’s hard to discern the actual proportion of in-frame: out-of-frame peptide responses. We also don’t know what rate of ribosomal frameshifting should be expected in the first place.
When comparing responses in humans the effect doesn’t seem as strong, although still somewhat noticeable.
Here, 21 vaccinees of mRNA vaccines and 20 vaccinees of adenoviral-based vaccines had their blood drawn and measured for IFNγ responses:
We then compared IFNγ ELISpot responses to predicted +1 frameshifted SARS-CoV-2 spike protein products in 21 individuals vaccinated with BNT162b2 and compared these responses to those of 20 individuals vaccinated with ChAdOx1 nCoV-19, none of whom reported undue effects as a result of vaccination.
Unfortunately, this is where problems start to arise and where media reporting goes off the rails.
Note here that the researchers never mention how many doses these individuals received. They also don’t provide any information on prior infection history based on what I’ve looked at.
This is important because the mice used in this study were given 3 doses of each vaccine. We can hypothesize that more doses would mean more chances for ribosomal frameshifting to occur, and so this would be one correlate that isn’t being looked at in this study. For instance, fewer doses of the mRNA vaccines may mean fewer out-of-frame peptide formation and therefore fewer immune responses to these frameshifted peptides. There’s also a possibility that increased exposure to the virus itself or the unmodified adenoviral-based vaccines may have a chance of increased ribosomal frameshifting (a speculation here).
And so we can’t really tell much about these participants aside from which vaccines they got. It’s rather strange that this study seems to lack any description of these patients that would be pertinent.
The results here are also not that impressive:
Similar to the mouse graphs the left shows IFNγ responses to frameshifted peptides while the right graph shows responses to in-frame peptides.
Relative to the mice models the responses here are far lower, although this could be due to the fact that blood samples were taken from vaccinated participants whereas the mice had their spleens removed and examined, so it’s possible that this may have led to the stronger response in mice due to more responsive cells.
But even with that being said, note that the responses in Pfizer humans are rather sporadic, appearing to be driven by 2-4 outliers in particular. Like with everything, the important question isn’t that some people show responses to frameshifted proteins, but why some people show these outlier-like responses.
Keep in mind that the sample size is also small for such a study, and given that no control was used we don’t know what the background rate to these frameshifted peptides should be. Remember that we don’t even know how many doses of each vaccine these participants were given.
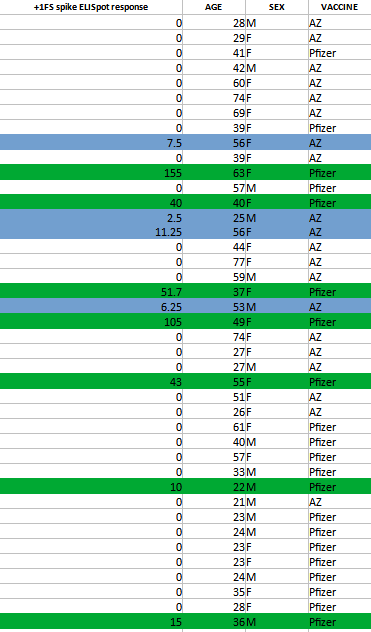
There’s also the fact that some of the AZ participants showed an IFNγ response. The overall breakdown can be found in the spreadsheet below with green noting the mRNA vaccinees and blue noting the AZ vaccinees who showed an IFNγ response:
To include or not to include…
At this point you may look at the AZ responses and consider them negligible. If that’s the case, I would rebut that argument with the following:
What immune response should we expect?
Let’s suppose that we should expect no response at all to these off-proteins.
If that’s the case, then we may allege that 7/21 Pfizer vaccinees and 4/20 AZ vaccinees show immune responses to off-proteins, or around 33% response vs 25% response, respectively. Would these be comparable? I’m not sure given the very small sample findings.
And if this seems like me being a pedant note that Science covers this study with the following remark:
Blood samples from about one-third of the Pfizer recipients showed an immune reaction to the frameshifted proteins, whereas none of the samples from AstraZeneca recipients did. None of the people reported any side effects from their particular vaccine, and there is no evidence, the researchers say, that the frameshifted protein is harmful.
It seems Science included all Pfizer participants who showed a response, meaning that this “no response” comment doesn’t quite work for AZ vaccinees. It’s obvious that some AZ vaccinees showed a response, so why report that none did?
It seems that reporting here may be doing a bit of number manipulation. It’s far easier to make things sound worse than they may appear when percentages are used, especially when sample sizes are as low as they are in this study.
One-third or 33% sounds rather scary. 7/21? Maybe not so much, and remember that this 7/21 is based on a sample size which we have no thorough information on, so we can’t even extrapolate this to larger populations.
Something that leads to nothing
The overarching problem with this study is that there is no way of interpreting the findings. How much of an immune response to off-proteins would be considered tolerable? What is the threshold, and what exactly is the background rate? Did people within the AZ produce an IFNγ response or not, and what measure would you use to make that argument to include or exclude?
And are these proteins harmful or not? I’ll break from both sides and say that this study and any inference from it can’t provide any comment for or against possible adverse reactions.
On one hand, the Science article and the researchers are makings claims that the off-proteins aren’t doing much:
None of the people reported any side effects from their particular vaccine, and there is no evidence, the researchers say, that the frameshifted protein is harmful.
No evidence from the researchers they say? And what evidence did the researchers provide to make that claim?
The only evidence I could find was the remark from the above excerpt which noted that none of the vaccinees reported “undue effects” due to the vaccines…and that’s about it- a highly subjective response from participants that they were OK after vaccination. Nowhere is additional information provided about these individuals medical background or of any tests conducted to confirm this assumption.
Remember that absence of evidence is not evidence of absence. Just because a participant says they didn’t notice any adverse effects does not mean something didn’t occur.
On the other hand, several people have taken to speculating that these off-proteins may be harmful. They are, after all, unintended products. But note that translation is not a 1:1 perfect process. Viruses appear to utilize frameshifting in order to produce some of their structural proteins, seemingly using this method as a form of cramming in more functional proteins into a smaller genome.
Crappy proteins are often made by the body as well- it’s usually when the threshold for too many misfolded or poorly-translated proteins is met that an issue may arise.
Again, the problem here isn’t whether these off-proteins are safe or harmful. We just don’t know because this study wasn’t designed to measure that, and any comment to the contrary would be mere speculation.
AND, if one were to argue that these off-proteins are harmful, it would have to rationalize the distinction between Pfizer/mRNA adverse events compared to adverse events found with the adenoviral-based and protein-based vaccines.
In short, this study can really be boiled down to the following:
Some people produce immune responses to off-target proteins. Which people? We don’t know. Maybe those given mRNA vaccines, but we don’t know how much is concerning or if other reasons may explain any outliers.
We don’t know what the response to these proteins, or what the proteins themselves mean since nothing was tested to show something one way or the other.
Personally, that’s all I can derive from this study.
Now, as to why this frameshifting may be occurring, the authors surmise that m1Ψ-rich codons may be causing the stalling as the ribosomes have difficulty parsing what codon they are actually reading. This seems to be supported by the fact that some of the off-protein products occurred at these m1Ψ-rich codons (Figure 4), thus suggesting that m1Ψ-rich codons within the mRNA vaccines may serve as slippery sequences.
If true, the results of this study are rather paradoxical to what we’ve been told about these vaccines. Note that one concern was that these mRNA vaccines have been codon optimized. Following the logic I mentioned about regarding rare tRNAs, codon optimization is when a codon for one amino acid is swapped for a different codon that still codes for that same amino acid. In this case, the swap is from a rarer tRNA to a more common one which may speed up translation, essentially removing possible ribosomal stalling as the ribosome waits for that rare tRNA. This has been alleged to produce possible off-proteins structurally, as speeding through translation may not allow room for proper folding of proteins during ribosomal stalling.
In contrast, if these m1Ψ-rich codons are themselves ribosome stallers then it seems as if the effect of codon optimization would be reversed. Ironically, the Science article themselves suggest that a correction would be to remove these m1Ψ-rich codons for ones that bear fewer m1Ψ:
But there’s a simple solution, the researchers say. Because mRNA can use several codons for a given amino acid, the molecules can be designed to avoid the slip-prone codons. For example, mRNA designers can use the sequence UUC (two pseudouridines and a cytidine) instead of UUU (three pseudouridines). Both code for the same amino acid, but the former should produce fewer errors. It’s a bit like writing mom instead of mum, Willis says. “It means the same thing.”
So in order to solve the problem of m1Ψ-rich codons you should… use less m1Ψ? Seems like creating a problem then offering a solution to said problem…which was just the original thing to begin with…
Anyways, I’ll leave readers with that. If anything needs clarifying please feel free to ask! Hopefully I can respond if not busy with Christmas and life-related things.
If you enjoyed this post and other works please consider supporting me through a paid Substack subscription or through my Ko-fi. Any bit helps, and it encourages independent creators and journalists such as myself to provide work outside of the mainstream narrative.

Mulroney, T.E., Pöyry, T., Yam-Puc, J.C. et al. N1-methylpseudouridylation of mRNA causes +1 ribosomal frameshifting. Nature (2023). https://doi.org/10.1038/s41586-023-06800-3











Why would you want anything to be produced inside your body triggered by a thing with which you were not born?
Analogous or not, irrelevant. It is completely irrelevant what this thing makes you to produce.
You are born as a 100% complete mechanism. Everything is in its place, doing its work, performing its role. No redundancy, no excess, no shortages. You are exactly as you should be.
Every cell of your body is 100% time occupied with what it does. No room for extracurricular activities.
And now you inject an “instruction” for ALL cells of your body to stop doing what they are doing and start producing something else. It is the maximum unimaginable stupidity, beyond any limits of fantasy. No matter how “scientifically” you name it, no matter how may “scientists” you will pay to “prove” the point. It’s way beyond stupidity - if you are not a believer, particularly.
If you declare believing in God, of any name, of any tradition - these injections, this mechanism, are a direct cancellation of your faith, of your religious beliefs, of the spiritual essence of your life. You are saying to God: “Your creation is defective. It doesn’t need what You did, our scientists know better than You.”
Add Dr Been to the list of folks covering this paper. He did a pretty good video last night on it.
https://www.youtube.com/live/BtiH5sfFklw?si=Ehl9HPQC2eQnFawd