Evidence of Molnupiravir-related SARS-COV2 mutations?
A recent study notes evidence of SARS-COV2 mutations possibly related to Molnupiravir, suggesting a link between viral mutations and the drug's use.
Molnupiravir has been one of the first drugs I have covered since starting this Substack. In my opinion, it’s one of the most contentious drugs used for the treatment of COVID due to its mechanism of action of being a mutagen.
Mutagens are inherently dangerous because they operate by inducing mutations within a pathogen’s genome. However, there’s also a possibility that these mutagens may be utilized by our own cells and therefore become harmful to us. There’s also the fact that mutagens don’t inherently destroy a virus, but gamble on the idea that a virus will accrue so many mutations that it no longer becomes viable; a phenomenon usually referred to as an “error catastrophe” (or something similar).
I have written a series on Molnupiravir detailing some of these events, although care should be taken to understand that some of the information may be outdated or may contain some errors due to the information available at the time.
Of note, one of the most striking situations was the FDA approval meeting where Molnupiravir was seen as such a contentious drug due to its mutagenesis that several members of the committee suggested that people who were prescribed Molnupiravir should be isolated from their family in case virions containing these mutations escape and infect others within a household.
Recently, several people such as Igor Chudov have noted that there appears to be evidence of escape mutations.
And that leads us to a few weeks ago where a preprint was published1 indicating a signal of mutations related to Molnupiravir use in sequencing databases.
I was alerted to this article by a friend, and Peter of All Facts Matter reported on this article just a few days ago as well.
In short, it appears that sequencing databases not only show a bias towards mutations expected from Molnupiravir, but that these mutations also appear to occur in places where Molnupiravir was widely distributed. Also, the evidence also suggests that transmission of these mutations to other people may be occurring, raising serious concerns over this drug’s widespread use.
Molnupiravir MOA and Relevant Terms
Since it’s been some time a few readers may need a refresher on how Molnupiravir works (this also adds more additional context that may have been missed in previous articles).
Molnupiravir comes from a class of drugs called nucleoside analogues. As the name of these drugs implies, their structures are analogous to our typical nucleosides such as adenosine, cytosine, uracil, thymine, and guanine, with slight modifications made to confer an antiviral mechanism.
A clear example of a nucleoside analogue is Remdesivir, which is an analogue of adenosine where the only modification is on the 1’ position of the sugar where a hydrogen (H) is replaced with a cyano (CN) group.

With Molnupiravir the main modification is done to the nitrogenous base. Usually such modifications alter the base pairing behavior of the nucleotide, and if mistaken for the wrong nucleotide a polymerase may incorporate a different base to hydrogen bond, thus leading to a drug-induced mutation.
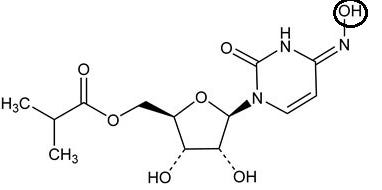
Molnupiravir only differs from cytosine by the addition of the hydroxyl (OH) group circled above. Since it’s an addition to the nitrogen on the 4th atom of the ring this leads to the name N-4-hydroxycytidine.
The addition of this hydroxyl group causes the active form of Molnupiravir to alternate between two different forms— an event called tautomerization. Here, the bonding pattern of the nitrogenous base may alternate such that the nitrogen that is part of the ring structure may gain or lose a hydrogen.
This can be seen in the structure in the bottom right of the following figure when comparing the “U” form and the “C” form:

Note that the “C” form listed above is recognized as being similar to typical cytosine while the “U” form may be recognized as a uracil.The gain and loss of a hydrogen is key as it alters whether the nitrogen will act as a hydrogen bond donor or a hydrogen bond acceptor. As a bond acceptor in the “C” form the base can properly pair with G (noted on the left of the image above). In the “U” form the nitrogen acts as a bond donor due to the N-H bond. As a bond donor the “U” form can pair with A (note that the ring of Adenosine shown in the bottom left image is missing a double bond and the nitrogen from N should not be bonded to a hydrogen as depicted in the image).
Molnupiravir’s effects aren’t direct. Instead, the drug needs to be incorporated into the virus’ genome during replication. It’s only during the second cycle of replication where the recognition of Molnupiravir as either a C or U becomes important.
*Remember back to biology and base pairing:
A base pairs with either a U or T
G base pairs with C
In this case, if Molnupiravir is recognized as a “C” during replication then the base may be incorporated opposite of a G in the growing viral strand. Afterwards, it may tautomerize into the “U” form. Following a second round of replication if the base is recognized as a “U” rather than the initial “C” because it tautomerized, then the viral polymerase will incorporate the wrong base (in this case an A rather than the correct G due to a recognition of a “U” form of Molnupiravir).
Because the wrong base was incorporated a mutation has occurred in the viral genome. In this case, the shift from a G to an A is referred to as a G-to-A mutation.
This is referenced often in the literature so it’s important to know what a G-to-A mutation refers to.
Interestingly, remember that Molnupiravir can be incorporated in either the C or U form, so two methods of mutagenesis is possible.
If instead Molnupiravir takes on the “U” form then it may be incorporated opposite an A. After it gets incorporated during the first round of replication, the “U” form of Molnupiravir may tautomerize into the “C” form. During the second round of replication if the viral polymerase recognizes the “C” form then a G will be incorporated. This mutation is considered an A-to-G mutation. It’s considered that this mutation should occur less often as it is less likely that Molnupiravir gets incorporated in its “U” form.

Note that G-to-A mutations are paired with C-to-U mutations. Although outside the scope of this review, be aware that single-stranded RNA viruses undergo more than one replication cycle, altering between what’s called a positive sense RNA and a negative sense RNA. What’s important to know is that incorporation of Molnupiravir during certain steps may cause either a G-to A mutation or a C-to-U mutation. The same goes for an A-to-G mutation, which is paired with a U-to-C mutation.
If this all sounds confusing, just remember that Molnupiravir’s mutagenic properties are derived not from its incorporation into the viral genome, but its incorporation and tautomerization into another form. It’s the switcheroo that is key. Also, remember that the key mutation to note is the G-to-A and C-to-T mutations in this study.
Lastly, the study refers to point mutations called transition mutations. Transition mutations are when one base is altered for a base of a similar category, such that a purine is swapped for another purine (i.e. A←>G) or a pyrimidine is swapped for another pyrimidine (C←>T←>U). This is in contrast to transversion mutations in which a purine/pyrimidine is altered for the other type of base. For instance, a swap from A→C would be considered a transversion mutation.
Here, all of the mutations in question are transition mutations.
Study Findings
When researchers examined recorded phylogenetic branches for SARS-COV2 they noted that many of the branches showed high rates of G-to-A transition mutations while transversions were relatively low. This ratio of G-to-A also appears to have increased in 2022 when Molnupiravir was distributed (Fig. 1A). Also, it appears that this set of mutations was different than that typically seen in SARS-COV2 mutations.
This high ratio of G-to-A mutations also appear to have occurred in countries with wider use of Molnupiravir such as the UK, US, and Australia (Fig. 1C). This is in contrast to countries such as Canada and France which prohibited the use of Molnupiravir.
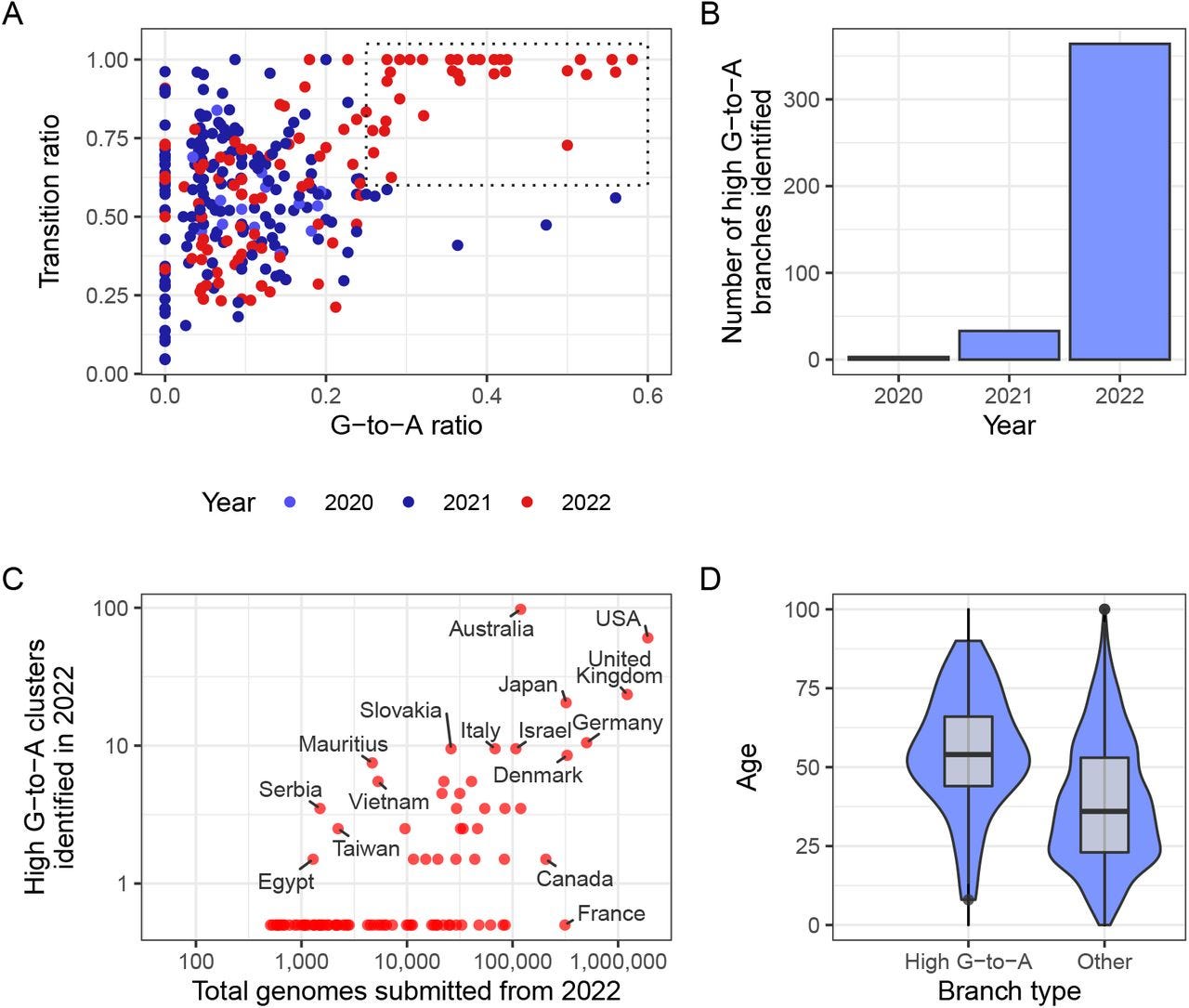
Note that clusters refer to samples from individuals with a shared phylogenetic branch.
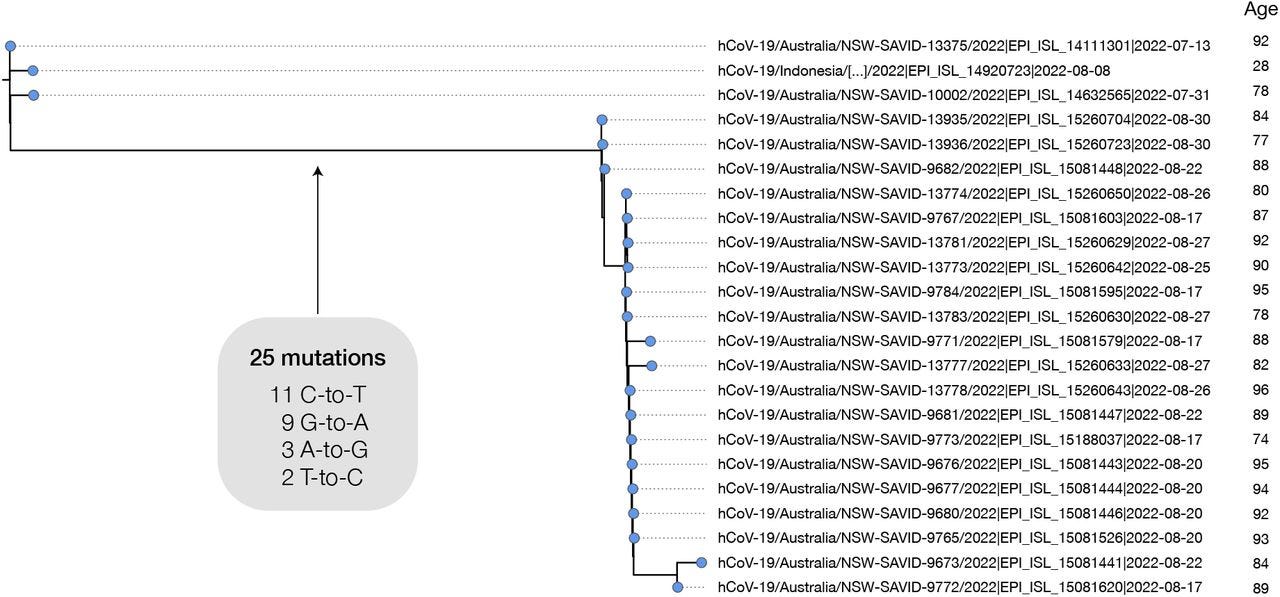
The researchers provide a case example from Australia in which 20 patients had 25 substitutions across the genomes isolated from these individuals individuals:
Most of the long branches sampled have just a single descendant tip sequence in sequencing databases, but in some cases branches have given rise to clusters with a significant number of descendant sequences. For example, a cluster in Australia in August 2022 involves 20 tip sequences, with distinct age metadata indicating that they truly derive from multiple individuals (Fig. 5). This cluster involves 25 substitutions in the main branch of which all are transitions, 44% are C-to-T and 36% are G-to-A. Closely related outgroups date from July 2022, suggesting that these mutations emerged in a period of 1-2 months. There are many other examples of high G-to-A branches with multiple descendant sequences, including from the United Kingdom (Fig. 6A,B).
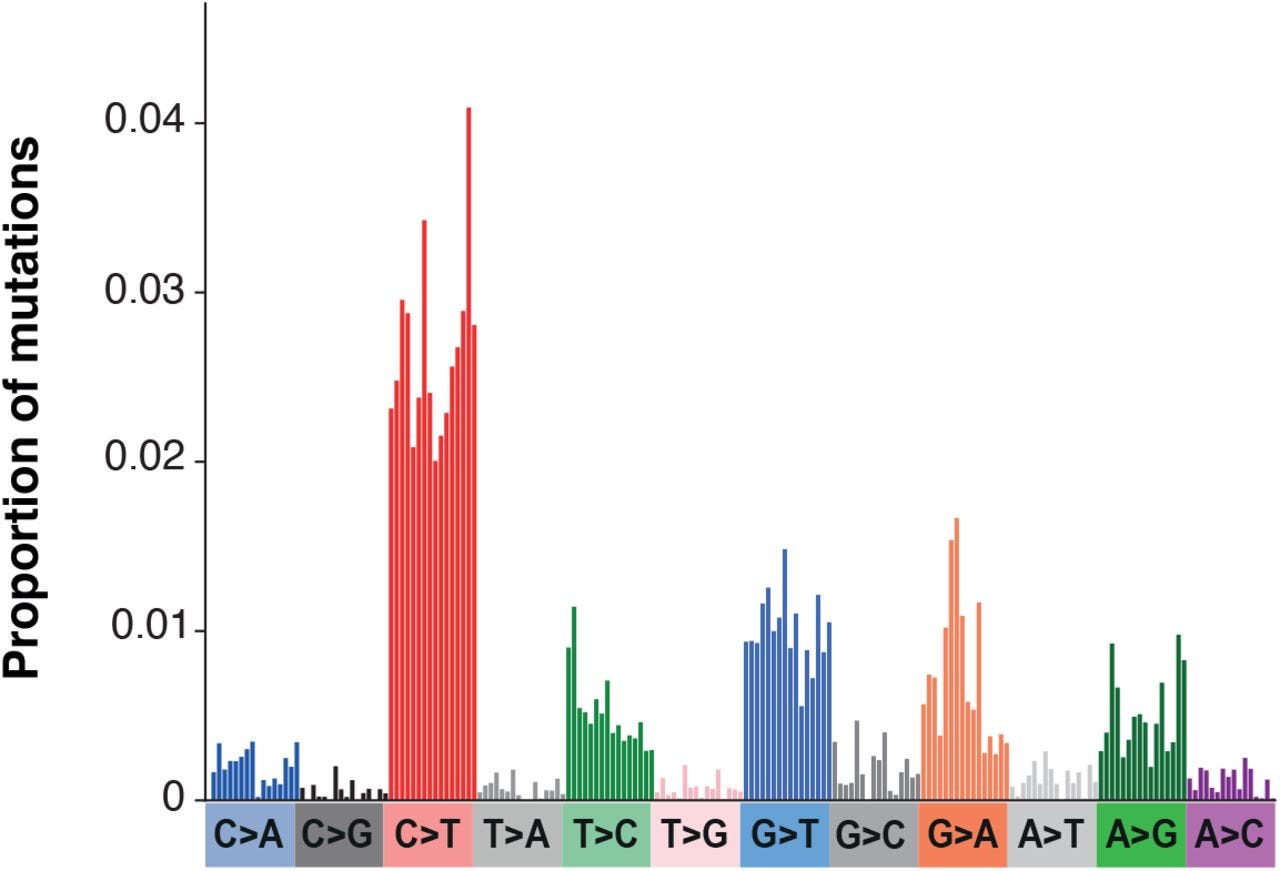
To further examine the extent of mutations the researchers created a mutational spectrum (frequencies of specific mutations) from global phylogenetic data and noticed a high degree of G-to-A/C-to-T mutations, with some A-to-G/T-to-C mutations also noted.
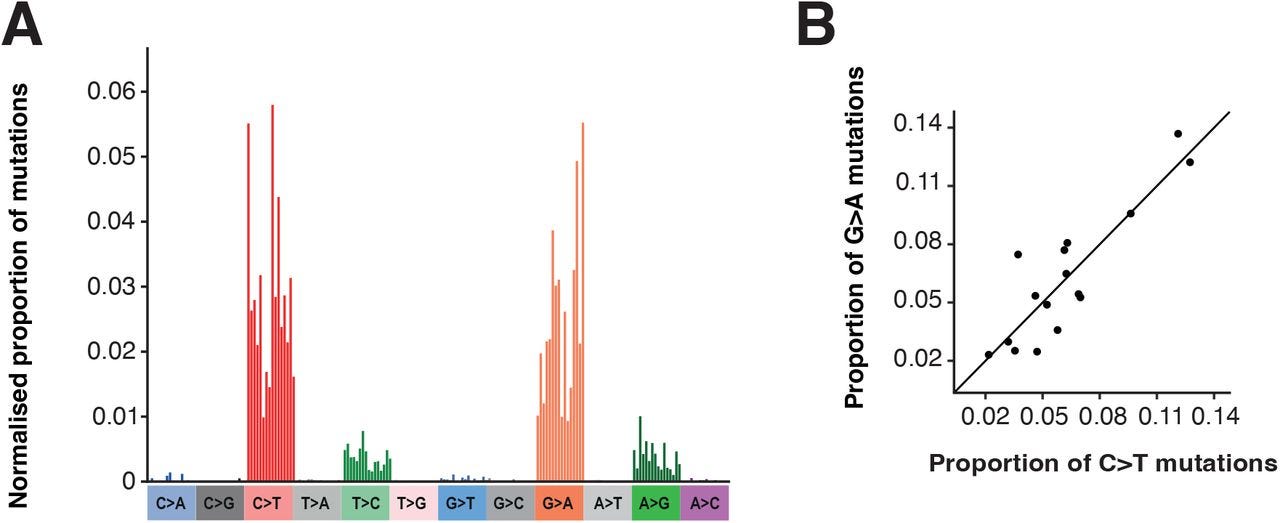
As noted above, this pattern appears similar to that of Molnupiravir’s mechanism of action.
However, because many of these mutations are speculated upon (i.e. they aren’t directly tied to actual Molnupiravir use) the researchers constructed a mutation spectrum with data from the AGILE Phase IIa clinical trial which provided mutation data on both the Molnupiravir and the control group.
Here, the data from the AGILE Phase IIa trial appear similar to that of the global phylogenetic data, although with a strange bias towards G-to-T transversions in both the control and Molnupiravir group not seen in the global mutational spectrum.
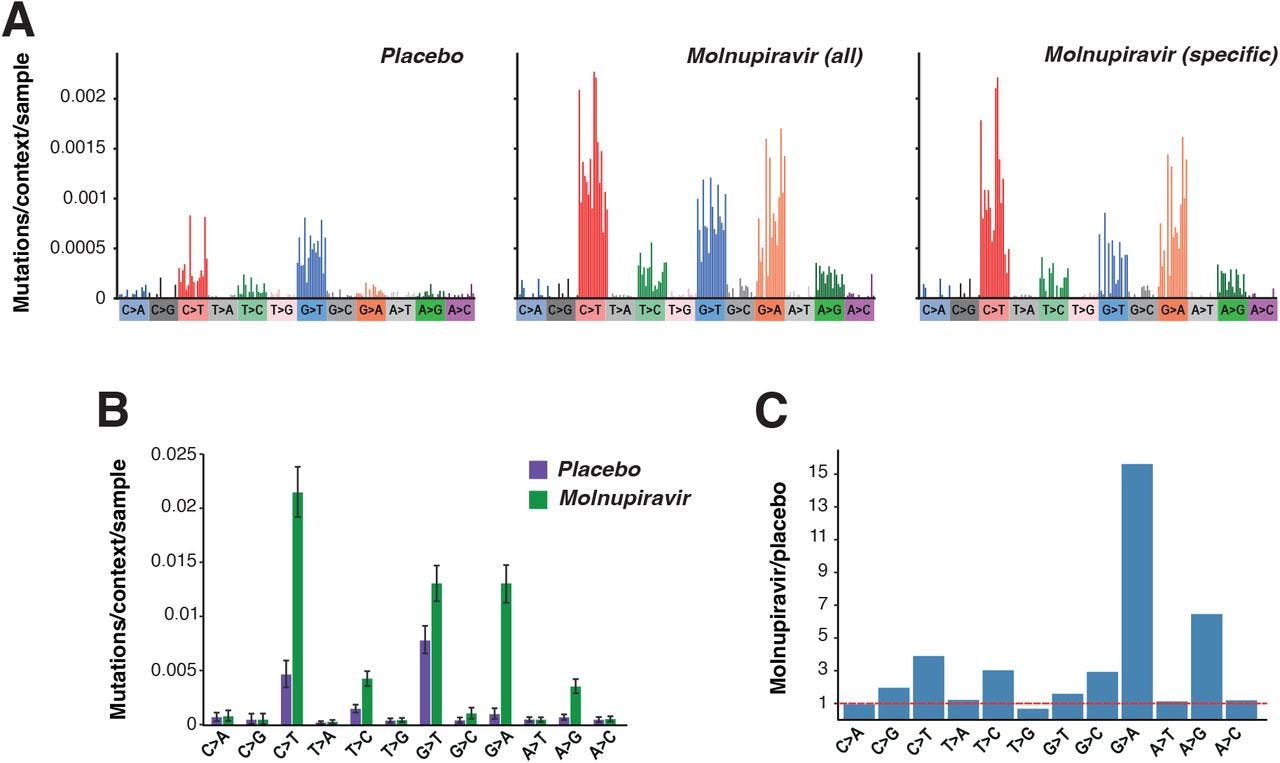
This may be due to difference in y-axis measurements between the global data, which appears to pool mutation data together and looks for frequency among the population, compared to the AGILE data which show spectra for each participant.
Given that those infected with SARS-COV2 are likely to be full of SARS-COV2 mutational soup it’s likely that the global data doesn’t capture the pool of mutations among individuals. Note that GISAID codes new sequences, and therefore would not measure prevalence of each mutation.
The use of proportions may also be masking the prevalence G-to-T transversions. As noted in Supplementary Figure 82 mutations in SARS-COV2 appears to show a high bias towards C-to-T and G-to-A mutations, as well as some of the mutations found in those given Molnupiravir.
It’s also possible that the difference in groups may be responsible for these differences as well, as the AGILE study included patients 18 and older while prescription of Molnupiravir appears to have been given predominately to older individuals:
Age metadata from the US showed a significant bias towards patients with older ages for these high G-to-A branches, compared to control branches with similar numbers of mutations but without selection on substitution-type (Fig. 3D). Where age data was available in Australia it also identified long branches primarily in an aged population. This is consistent with the prioritised use of molnupiravir to treat older individuals, who are at greater risk from severe infection, in these countries. In Australia, molnupiravir was pre-placed in agedcare facilities, and it was recommended that it be considered for all patients aged 70 or older, with or without symptoms (Australian Department of Health and Aged Care, 2022).
Concluding Remarks
Overall, what this study appears to suggest is a high bias towards mutations one would expect in a population given Molnupiravir, suggesting that the drug may be driving some of the mutations being recorded.
In the case of the specific cluster of 20 individuals the age of the patients would suggest that the data should be interpreted with immunosenescence in mind. That is, that many of these patients likely have poor immune function and thus would not be able to clear the virus on their own. It’s possible that the use of Molnupiravir in this group may not lead to proper viral clearance, thus allowing for selective variants to spread among a vulnerable population (if these people were provided Molnupiravir). This effect has been seen in other data on Molnupiravir, such as the Fountain-Jones, et al.3 data in which the researchers note the possible risk of Molnupiravir for immunocompromised individuals given the possibility of unchecked mutations.
Sanderson, et al. note that some of these sequencing data may be related to chronic infection in immunocompromised individuals:
The cases we identified with these very high numbers of mutations involved single sequences, and could represent sequences resulting from chronically infected individuals who have been treated with multiple courses of molnupiravir.
So this raises questions as to whether the drive seen may be due to the cohort that Molnupiravir is prescribed to i.e. the elderly, who are not able to properly clear the virus with their own immune systems, thus serving as reservoirs for many variants.
So, is this study a slam dunk in Molnupiravir driving mutations?
I’d suggest that it adds a lot more to the concerns over the drug, and it suggests that further research should be done to examine case reports in those who are immunocompetent to see if mutations may arise in these individuals as well.
It would appear that these events may be confounded by age and immunity status, which require further considerations given that these groups are the ones likely to be prescribed Molnupiravir.
Also, remember that the significance of these mutations were not noted. Although many mutations appear to have accrued, there isn’t any information with respect to the boost in virulence or whether these mutations were actually deleterious to the virus in some ways.
GISAID data also notes new sequences found, but doesn’t tell us anything about the prevalence. It’s possible that these new sequences may have been detected but extremely low in prevalence among the group it was collected from.
With all that being said, given that Molnupiravir was approved under the most contentious of circumstances the mounting evidence raises questions as to whether this drug should continue to be prescribed.
As described by the researchers, Molnupiravir is a gambling drug, in that one assumes that the virus accrues enough mutations that it ceases to function, but that doesn’t mean that will actually happen:
Molnupiravir’s mode of action is often described using the term “ error catastrophe” – the concept that there is an upper limit on the mutation rate of a virus beyond which it is unable to maintain self-identity (Eigen, 1971). This model has been criticised on its own terms (Summers and Litwin, 2006), but is particularly problematic in the case of molnupiravir treatment. The model assumes a steady-state condition, with the mutation rate fixed at a particular level. The threshold for error catastrophe is the mutation rate at which, according to the model, the starting sequence will ultimately be lost after an infinite time at this steady-state. However molnupiravir treatment does not involve a steady state, but a temporarily elevated mutation rate over a short treatment period. Therefore there isn’t any particular threshold point, or well-defined “ catastrophe” condition. We suggest that the use of this term in the context of mutagenic antiviral drugs is unhelpful. It is enough to think of these drugs as acting through mutagenesis to reduce the number of viable progeny that each virion is able to produce – particularly given that much of the reduction in fitness will be due to “ single-hit” lethal events (Summers and Litwin, 2006).
Put another way, the mutations may accrue but the catastrophic moment may never arise, and so the use of “error catastrophe” is a bit of a misnomer as it assumes an event that may not happen.
Right now China is utilizing Molnupiravir to deal with their COVID wave, and Merck has come out against the study suggesting that it only provides “circumstantial” evidence of the association between Molnupiravir and these mutations.
Ironic, given that Molnupiravir was approved under circumstantial evidence, and yet it’s that evidence that is now being questioned.
Identification of a molnupiravir-associated mutational signature in SARS-CoV-2 sequencing databases
Theo Sanderson, Ryan Hisner, I’ah Donovan-Banfield, Thomas Peacock, Christopher Ruis
medRxiv 2023.01.26.23284998; doi: https://doi.org/10.1101/2023.01.26.23284998
Antiviral treatments lead to the rapid accrual of hundreds of SARS-CoV-2 mutations in immunocompromised patients
Nicholas M. Fountain-Jones, Robert Vanhaeften, Jan Williamson, Janelle Maskell, I-Ly J Chua, Michael Charleston, Louise Cooley
medRxiv 2022.12.21.22283811; doi: https://doi.org/10.1101/2022.12.21.22283811




Wow, interesting, I didn't expect that. I guess I lumped antivirals together, and knew that hydroxychloroquine and ivermectin were widely used. They must work differently than Mol.