The Curious Emergence of Omicron
Part IV: Attenuated Lung Infection, Change in Disease Pathology?
Omicron has Attenuated Targeting of the Lungs, Possibly Becoming more of an Upper Respiratory Infection
At the start of the pandemic initial reports of COVID symptoms usually detailed symptoms such as a cough, sore throat, and fever. However, the cough was usually described as a dry cough, consistent with a lower respiratory infection. Because of this, COVID infection was described as being an infection of the lower respiratory tract rather than an upper respiratory infection akin to seasonal cold and flu infections.
Of course, we now know that COVID can take on various disease pathologies, and in many circles have been considered a vascular disease due to the presence of clots in severe, late stages of the disease.
Omicron has presented itself with a much different route of infection compared to its viral counterparts, and one would have to wonder if these differences may lead to differences in disease presentation.
Indeed, emerging evidence suggests that Omicron has reduced infectivity of cells of the lung, which may be a signal of a move towards an upper respiratory pathogen.
In short, the respiratory tract, which includes the nose, throat and lungs can be broken up into two regions. The upper respiratory tract consists of the nose, pharynx, and larynx (the larynx seems to be a point of contention for which “side of the tract” it belongs) and usually ends near the top of the throat. The lower respiratory tract, on the other hand, involves the trachea, bronchi, and lungs.
If true, this would present with a startling change in disease pathology, as it would add weight to the idea that Omicron is a much more mild form of COVID; a position that would provide a great deal of optimism for the world at large considering how quickly it is moving through the population.
In one final reference to Zeng et. al., researchers combined Omicron pseudo lentiviruses with human lung-epithelial derived CaLu-3 cells. Fluorescence measures were taking (higher binding to lung cells equated to higher fluorescence) and Omicron showed a far lower level of fluorescence, suggesting a reduction in infectivity towards human lung epithelial cells compared to prior circulating strains of SARS-COV2.

Careful considerations should be taken with this study, especially after our extensive commentary on Omicron’s heavy dependency on cell-to-cell transmission, which would not be picked up in this assay. Nonetheless, it adds evidence to the argument that Omicron is less lethal and virulent.
Similar results were obtained in an ex vivo assay by Chan et. al., in which researchers infected human bronchus and lung cells with the D614G, Delta, and Omicron strains of SARS-COV2. Omicron showed far higher infectivity of bronchus cells but showed reduced infectivity of lung cells.

There’s a few things to point out here. Specifically, Omicron appears to infect bronchus cells very rapidly, with a high level of infection occuring 24 hours post-infection (hpi) and gradually increasing for the next 24-48 hour period. Also, Delta appears to have a slower start to its infection, although eventually reaching a higher level of infection 72 hours hpi compared to Omicron. More interestingly, Omicron doesn’t appear to have the same level of lung infectivity as compared to either D614G or Delta, and even seems to lag behind in its infectivity.
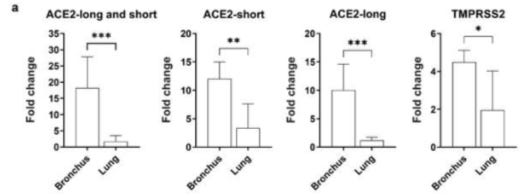
When WT, Delta and Omicron variants were compared, Omicron variant replicated to significantly higher titres than WT or Delta at 24 and 48 hpi; the difference being over 70-fold (Figure 1b). At 72 hpi, both Delta and Omicron viruses replicated significantly more than WT in human bronchus but there was no significant difference between Delta and Omicron viruses. When these experiments were done at 33oC, the viral titres were similar to those at 37oC for each virus (data not shown). In ex vivo cultures of human lung, the only significant difference observed was reduced replication of Omicron compared to WT at 24, 48 and 72 hpi (Figure 1c,d). These findings in bronchus and lung were confirmed in area-under-curve (AUC) analysis of aggregate virus titres at 24-72 hpi (Figure 1e,f).
The researchers suggest that this difference could be attributed to differences in ACEII and serine protease expression between bronchus and lung cells, such that bronchus tissue contains greater expressions of both proteins. However, it’s important to note that the dependence on both serine protease and ACEII expression seem to be reduced in Omicron, and the researchers suggested that serine protease may not play a significant role in Omicron (something also detailed in the Zeng et. al. study).
In the realm of lung tissue, these results are promising. They suggest that lung infection may be attenuated with Omicron, although not fully eliminated. These results do, however, throw a wrench into the idea of Omicron being a predominately upper respiratory infection, as the bronchi are in close proximity to lung tissue (as indicated by the anatomy diagram above). This raises questions as to the prior strains of the disease and possible dependency on lung infection for disease severity.
So far there is consistent evidence in support of reduced lung infection, but in order to determine the extent of the cells targeted by Omicron, ex vivo assays of nasal epithelium and cells of the throat would help to paint a better picture as to which cells are likely to be more targeted by Omicron.
Because using human subjects may be considered extremely unethical, animal models may be the best source of mimicking a human infection with Omicron. Luckily, there have been some studies utilizing hamster and mouse models as parallels to human infection.
A study by Diamond et. al. examined the effects of Omicron infection on mouse and hamster models. The researchers utilized various types of hamsters and mice, but for our sake we will look specifically at rodents transgenic for human ACEII expression.
Transgenic animals are animals genetically engineered to express genes from other sources. In this case, these rodents were engineered to express human ACEII receptors, and thus serve as good candidates to model human infection, although caveats must be taken into consideration on account for the differences in models, and thus serve more as simulacrums rather than as true parallels.
In the mouse study, the researchers inoculated transgenic mice with Omicron. The researchers noted that these mice did not exhibit weight loss (weight loss is considered a measure of disease severity in animal models), which counters previous results using wildtype/D614G which showed a reduction in weight loss in those studies.
Given the attenuation in several strains of conventional laboratory mice, two groups tested B.1.1.529 infection in 5 to 6-month-old (M.S.D. and A.G.S.) K18-hACE2 transgenic mice, which express hACE2 under an epithelial cytokeratin promoter12, and are more susceptible to SARS-CoV-2 infection and disease14. At intranasal inoculating doses ranging from 103 to 105 infectious units of B.1.1.529, weight loss was not observed over the first 5 to 6 days of infection in younger or older K18-hACE2 mice (Fig 1f). These data contrasts with historical results with WA1/2020 D614G or variant (e.g., B.1.351) SARS-CoV-2 strains14,22,34,36, which uniformly induce weight loss starting at 4 dpi. Although we observed clinical attenuation of B.1.1.529 in K18-hACE2 mice, the virus nonetheless accumulated in the upper and lower respiratory tract (Fig 1g).

What’s interesting here is that, contrary to the studies using human lung epithelial, these in vivo models show very high viral load in the lungs of these mice. It poses a few questions, as the researchers noted that a few mutations in the spike protein may tailor Omicron towards better binding to murine ACEII. If hACEII transgenic mice still express murine ACEII receptors, there’s a possibility that those receptors may play a tangential role in mice infection not seen in human cells.
Regardless, the results here are in line with the previously listed studies, as even in mouse models disease severity seems to be attenuated.
And the same can be said for hamsters expressing hACEII where these hamsters also did not show a reduction in weight compared to previous studies using wildtype virus.
Although hamsters are susceptible to SARS-CoV-2 infection without a requirement for host adaptation and show some similarities to that observed in COVID-19 patients, they develop self-limiting clinical and respiratory disease. Even though hamster ACE2 can serve as a receptor for SARS-CoV-2 spike protein, some of the contact residues in human ACE2 are not conserved38, which could diminish infectivity. Moreover, ACE2 expression levels on particular cells in the respiratory tract may differ slightly between hamsters and humans, which could impact infectivity and clinical outcome. To develop a more susceptible hamster model, members of the consortium (Y.K.) used transgenic hamsters (generated by Z.W.) expressing human ACE2 under the epithelial cytokeratin-18 promoter39. Whereas intranasal inoculation of 103 PFU of HP-095 D614G virus resulted in marked weight loss within the first week (Fig 2l) and uniform mortality by 10 dpi (Fig 2m), less weight loss and death (P < 0.05) were observed after infection with 103 PFU of B.1.1.529. Consistent with these clinical data, 1,000 to 10,000-fold lower levels of infectious virus were measured in the lungs of hACE2 transgenic hamsters challenged with B.1.1.529 compared to the HP-095 D614G virus at 3 and 5 dpi (Fig 2n). Notably, and as seen in wild-type Syrian hamsters, smaller differences in infection were observed in the nasal turbinates. Thus, B.1.1.529 infection in the lung is attenuated in both wild-type and hACE2 transgenic hamsters.

However, in the case of the hamster model, viral load in the lungs did not seem to differ greatly with the addition of hACEII. Several factors could account for this, such as number of animal models, measurements taken at different dpi’s, different calculations, and different Omicron strain (wildtype mice and hamsters were given a strain from Japan while the animals that expressed hACEII were given a strain from the US). All of these play confounding factors, but consistency between the wildtype and hACEII hamster models may suggest that hACEII may not play a big role in infectivity for Omicron, consistent with the Zeng et. al. study.
But that doesn’t quite explain why the humanized mice models showed such high viral loads in the lungs. It could be due to the use of different animal models and differences between murine and hamster ACEII expression and structure that have affected the results. Considering that many of Omicron’s mutations appear to confer enhanced binding to murine ACEII, and the uncertainty of binding dynamics between the spike protein and hamster ACEII, there could be a selection bias in favor of increased binding towards mice by virtue of the mutations.
If the humanized mice models still express murine ACEII as well as hACEII, there may be a synergistic effect such that both expressions may increase viral infection of the lungs. Again, there is a difference in time measurements within the mouse study that could factor in, and all of this is based on speculation.
Humanized animal models are not perfect representations of human pathogenesis, but it provides a window into the possible infection pathways that may occur in humans. Here, we’ve seen plenty of instances that suggest Omicron exhibits attenuated infection of the lungs, a model that correlates with real-world evidence (although preliminary as of now) of more mild illness. Although attenuated, it is not completely ablated so there is a possibility of severe infection among certain demographics.
There also lies the evidence of high viral load in the bronchus, and some inconsistent evidence of nasal viral load in the animal models. More studies would help to provide more consistent evidence of the effects of viral load in the upper airway and symptom presentation. Nonetheless, most reporting of Omicron has consisted of cold and flu-like symptoms, all suggestive of a possible move more towards an upper respiratory infection.
At the same time we’re piecing information together, there are plenty of questions left unanswered. Indeed, mounting evidence suggests that Omicron behaves extremely differently compared to it’s other counterparts, and it’s emergence and sudden, rapid global spread is reminiscent of the first emergence of COVID. In a similar manner, this raises questions on the possible origins of Omicron. Although we may speculate on this matter, there’s considerable evidence of a possible animal reservoir, in this case the possibility that mice may have served as a viral reservoir, and one in which Omicron may have emerged from.
Part V will look into the widely circulating study suggesting mice may be the origins of Omicron.




Love your in depth explanations of all things covid. Thank you so much.
Literally, the best analysis out there for us Ronanerds. Thank you for all your hard work.