Have Monoclonal Antibodies Lost their Effectiveness? (Review)
Part II: Examining Molecular Modeling and Nonclinical Studies
Molecular Modeling and Monoclonals
Molecular modeling can be a touchy subject. If nonclinical, in vitro assays raise concerns about the actual real-world, human effects of therapeutics then molecular models should garner the most scrutiny, as their predictive models depend on tons of variables that may be manipulated by their modeler and introduce plenty of bias into the results. Nonetheless, molecular models provide some of the best evidence of molecular dynamics, many of which are not easily captured by commonly used assays.
The emergence of Omicron raises questions of whether the monoclonals in circulation will lose effectiveness, and the best place to start in order to find evidence will be to examine the molecular dynamics of antibody/Omicron binding.
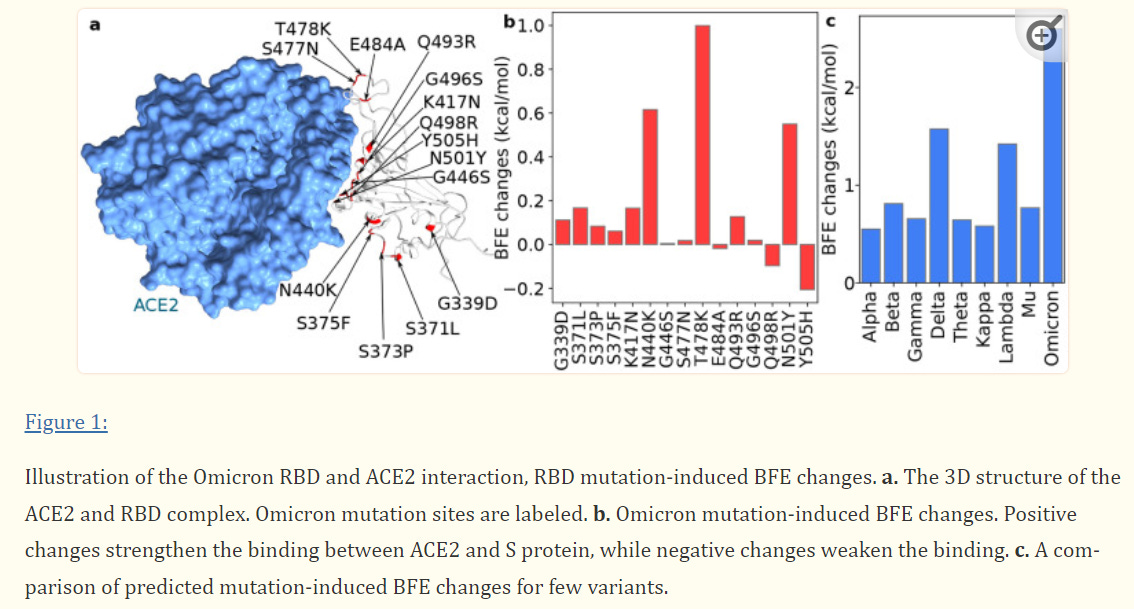
One of the only available modeling evidence at the time of this writing comes from a preprint by Chen et. al. in which researchers examined the effects of RBD mutations on binding free energy (BFE).
Binding dynamics depend on the thermodynamic nature of molecular interactions; favorable interactions lead to an increase in the release of free energy (exothermic interactions) while unfavorable interactions require outside energy in order to overcome the energy barrier (endothermic reactions). For the sake of the figures I included, know that BFE bars above the “0 mark” are considered favorable interactions (greater binding affinity) while those below the “0 mark” are considered unfavorable interactions (reduced binding affinity).
To start off, below is a figure indicating the BFE associated with Omicron/ACEII receptor binding. As should be expected, many of Omicron’s mutations provide greater binding affinity to the ACEII receptor, which is likely to play a role in its greater infectivity rate as compared to Delta. The researchers’ models suggest that the R0 (the infection rate) of Omicron could be around 10, nearly double that of Delta.
Eli Lilly’s Monoclonals Lose all Neutralizing Capabilities
Of all the circulating monoclonals, the dual therapy from Eli Lilly’s are the most likely to lose effectiveness due to the overlapping binding regions. As is expected, molecular modeling suggests that Eli Lilly will lose all neutralizing capabilities, likely due to the extremely large BFE change from Omicron’s mutations.
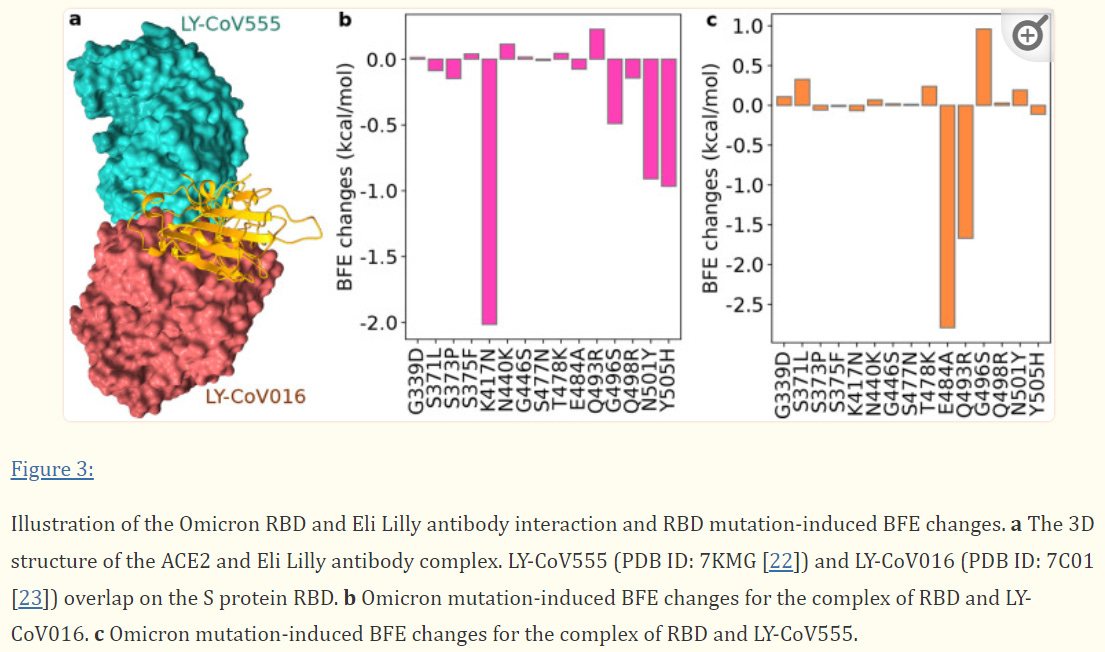
The researchers noted:
Although LY-CoV555 and LY-CoV016 might slightly complement, they are both prone to Omicron mutation-induced efficacy reduction. We predict that the Eli Lilly mAb cocktail would be retaken off the market had Omicron become a prevailing variant in the world.
What’s interesting is that many of these mutations (N501Y, E484A) have been found in prior variants, which adds evidence to the revoking of Bamlanivimab’s EUA due to reduce effectiveness. Eli Lilly’s dual therapy were optimized for the wildtype spike protein meaning that they were the most likely to become susceptible to any type of deviation from the original viral protein, and we can see that with the drastic loss of binding affinity for both monoclonals.
Because of this, Eli Lilly’s monoclonals are very likely to not prove effective against Omicron based on these preliminary data.
Regeneron’s Dual Therapy Likely to see Minor Reductions in Binding Affinity
When it comes to Regeneron the evidence isn’t as clear cut. Where some of the mutations may cause a loss of binding affinity, others appear to increase the binding affinity.
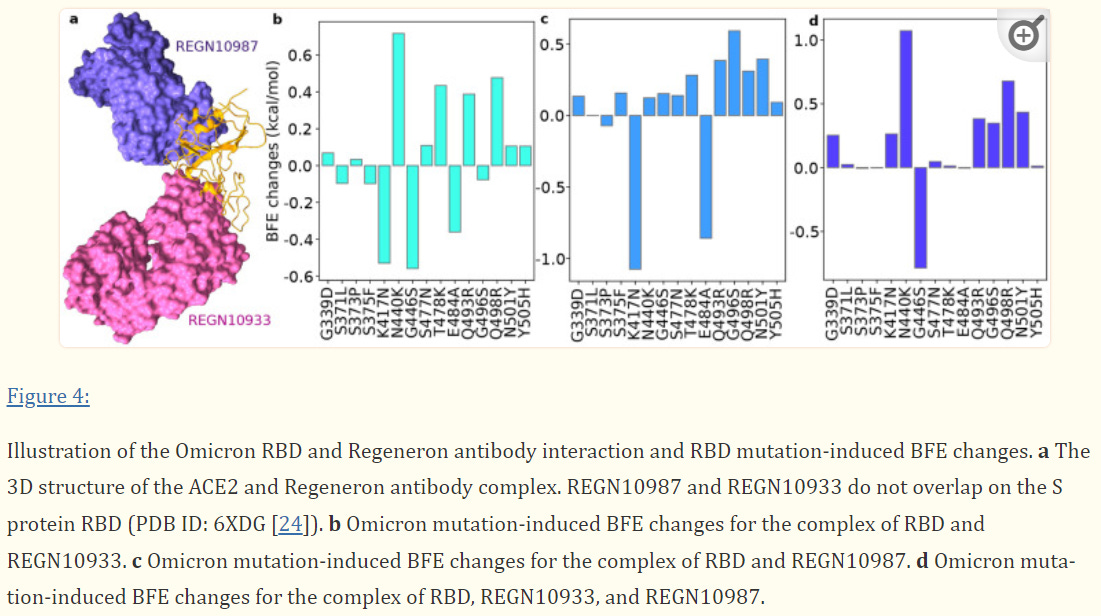
In the case of both Eli Lilly and Regeneron, the K417N mutation appears to lead to the greatest loss of binding affinity. This is a shift from a positively charged lysine (K) to a polar, uncharged asparagine (N) residue. This would suggest that the ionic interaction between the monoclonal’s paratopes and the lysine residue of the spike protein play a vital role in their neutralizing abilities.
This creates a difficult scenario. We’d like to believe that binding affinities are based on the “sum of its parts”, such that if the net strengthening binding affinities from mutations outweigh the weakening affinities, we may assume that the neutralizing effects of the monoclonals will hold up. In practice, this is more difficult to assess. For instance, the K417N mutation may prove so deleterious that it may prevent the binding of any of the antibodies, even if some of these mutations may actually strengthen the binding. In such a case, it does not matter whether binding affinity is strengthened if one deleterious mutation is enough to prevent binding.
The researchers suggest that Regeneron should see a minimal reduction in effectiveness, although many questions remain unanswered, and real-world evidence would be required to substantiate the claim raised.
Unfortunately, the researchers did not provide molecular modeling for Sotrovimab, likely due to Sotrovimab not targeting the ACEII binding region of the spike protein. Nonetheless, the researchers’ results suggest that Eli Lilly’s monoclonals should lose all neutralizing abilities while the deleterious mutations should leave Regeneron relatively unscathed.
This raises a good bit of skepticism about these results, especially in regards to Regeneron. The researchers have remarked that their prior models have garnered consistent results so it adds some leverage to the proposed models outlined above. However until more evidence, especially in clinical settings, are revealed these models are nothing more than predictive.
Unfortunately, emerging nonclinical studies are beginning to call into question the effectiveness of Regeneron as well.
Monoclonals do not Fare Well in Nonclinical Trials
Just like molecular modeling studies, in vitro studies comparing monoclonals against Omicron infection are very limited. However, the limited studies available show very concerning results.
First, it’s important to point out that the in vitro studies we will be looking at may not be representative of real-world scenarios. Remember that monoclonals have the capabilities of eliciting their own immune responses through effector-mediated functions, which won’t be picked up in assays that lack any immune cells. Also, several of these studies utilized pseudotype viruses or cells aside from normal human cells, meaning that the virus/cell binding dynamics may not be consistent with what would occur in humans infected with Omicron. It’s also worth noting that nearly all of the studies provided here are preprints and are subject to change after peer review and editing. Still, they provide a direction for further studies and possible clinical evidence.
In one study (Wilhelm et. al.) researchers isolated antibodies from previously vaccinated individuals and challenged Omicron variant SARS-COV2 virus particles. The researchers also examined Regeneron’s effects on neutralizing Omicron. Although both groups were effective at neutralizing Delta, there was great reduction found in Omicron neutralization for both classes of antibodies. More important to this newsletter, the study suggests that Regeneron’s inhibition capabilities were severely reduced.
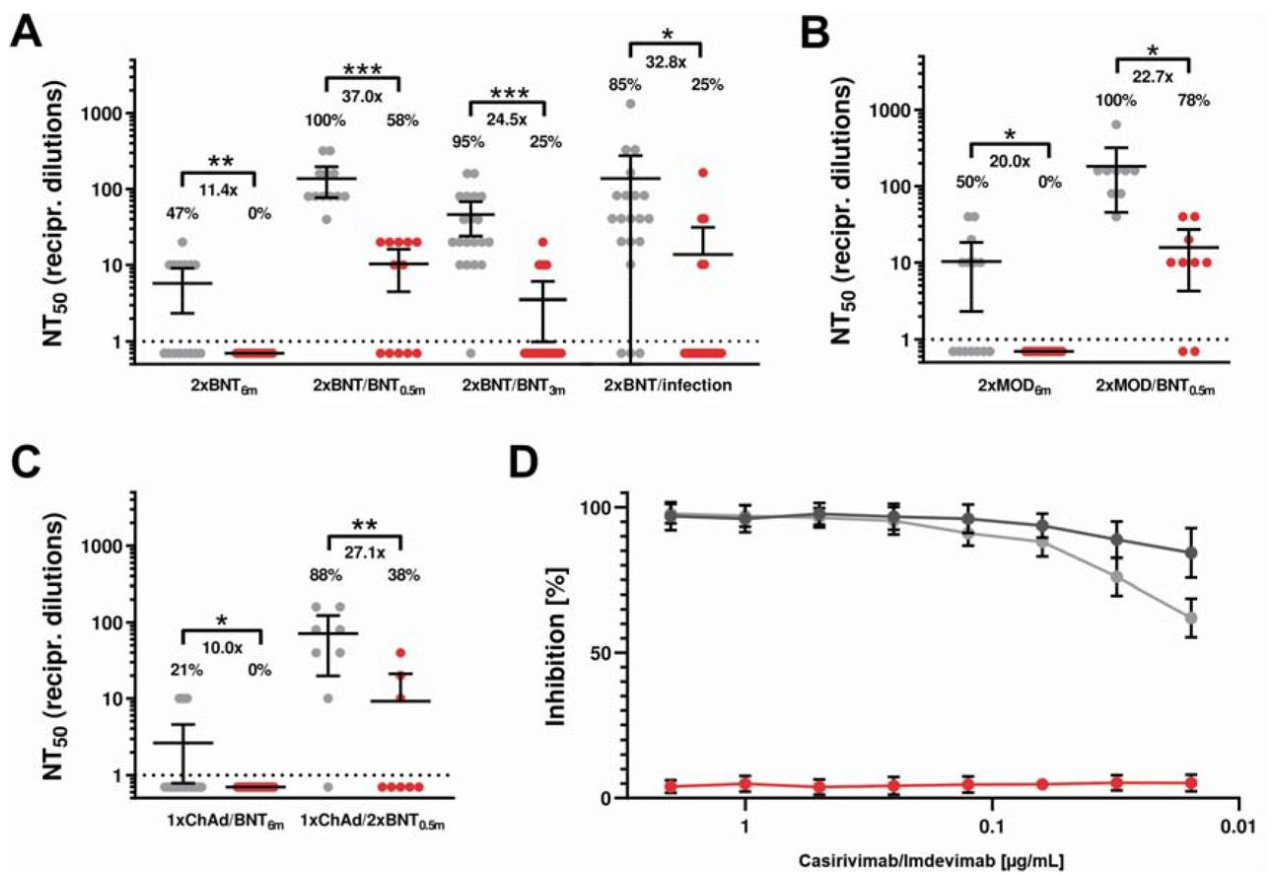
This raises an important concern. Although Omicron is gaining in prevalence Delta still makes up a good portion of infections. More importantly, if Omicron is believed to be far more mild than Delta we can expect that most COVID hospitalizations are likely to be Delta rather than Omicron. This begs the question; even if monoclonal antibodies such as Regeneron prove ineffective against Omicron, is it still important to provide monoclonals as early treatment in the off chance that the infection could be Delta?
In another preprint (Cao et. al.) researchers incubated Omicron pseudotype viruses with dilutions of therapeutic monoclonals. These viruses were then incubated with cells and the inhibition capacities were measured. Again, most monoclonals, including Eli Lilly’s and Regeneron’s, had drastically reduced inhibition capacities.
Some of the newly tested monoclonals, such as BRII which is undergoing clinical trials, and AstraZeneca’s AZD monoclonals unfortunately showed greatly reduced inhibition against Omicron, although not to the same levels as both Eli Lily or Regeneron’s. As expected, Sotrovimab (VIR-7831) showed minimal loss of inhibition (around a 2.5-fold decrease as compared to Delta), likely attributed to Sotrovimab’s targeting of conserved spike protein regions.
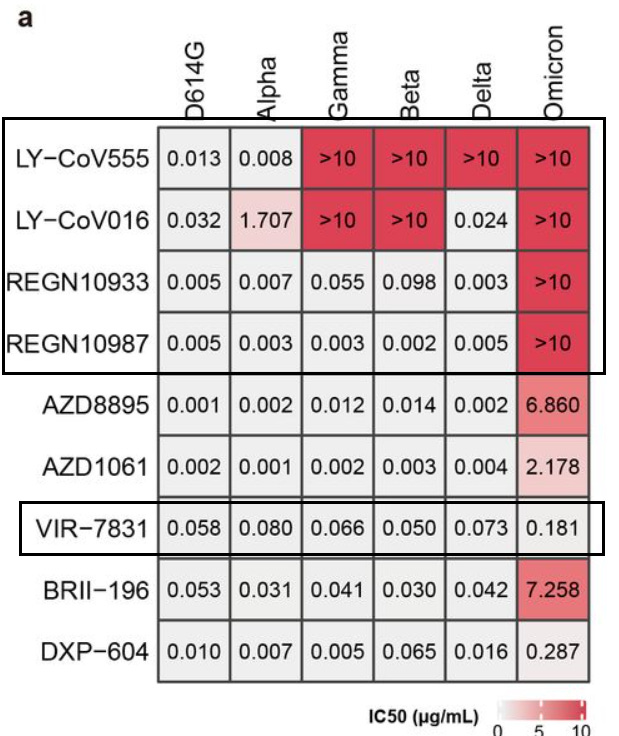
And as the researchers noted:
As for NAb drugs, consistent with their escaping mutation profiles, the neutralization potency of LY-CoV016/LY-CoV555, REGN-10933/REGN-10987, and AZD1061 are greatly reduced by Omicron (Fig. 4a, Extended Data Fig. 9). The binding affinity of AZD8895 and BRII-196 toward Omicron RBD is also significantly reduced, likely due to multiple mutations accumulating on their epitopes, such that AZD8895 and BRII-196 failed to neutralize Omicron (Extended Data Fig. 10). BRII-198 was not tested since the antibody sequence was not released. VIR-7831 retains strong RBD binding capability, although G339 is part of its epitope, the G339D mutation in Omicron does not appear to affect VIR-7831’s binding; however, VIR-7831’s IC50 is reduced to 181 ng/mL, and may be subject to further reduction against Omicron with R346K.
As an aside, it’s worth nothing tha Eli Lilly’s Bamlanivimab (LY-CoV555) had drastically reduce effectiveness against both Gamma and Beta variants, leading to the introduction of Etesevimab (LY-CoV016) as discussed previously. We can see that the effectiveness of this dual therapy against Delta is predominately derived from Etesevimab alone, indicating another fault with the developmental process of Eli Lilly’s monoclonal therapies. Take into account that Bamlanivimab’s effectiveness in this pandemic was very short lived as minor mutations led to its sudden lack of effectiveness. On this front, these nonclinical results are consistent with real-world evidence of these monoclonals’ effectiveness (or lack thereof) against prior variants.
Similar results were obtained by VanBlargan et. al. in which Vero-TMPRRS2 cells (cells expressing the serine protease required to cleave the spike protein’s furin cleavage site) and Vero-hACE2-TMPRRS2 cells (cells expressing both the serine protease as well as the human ACEII receptor required for viral binding). Once again, Sotrovimab was able to reduce viral infection while both Eli Lilly and Regeneron were unable to prevent infection.
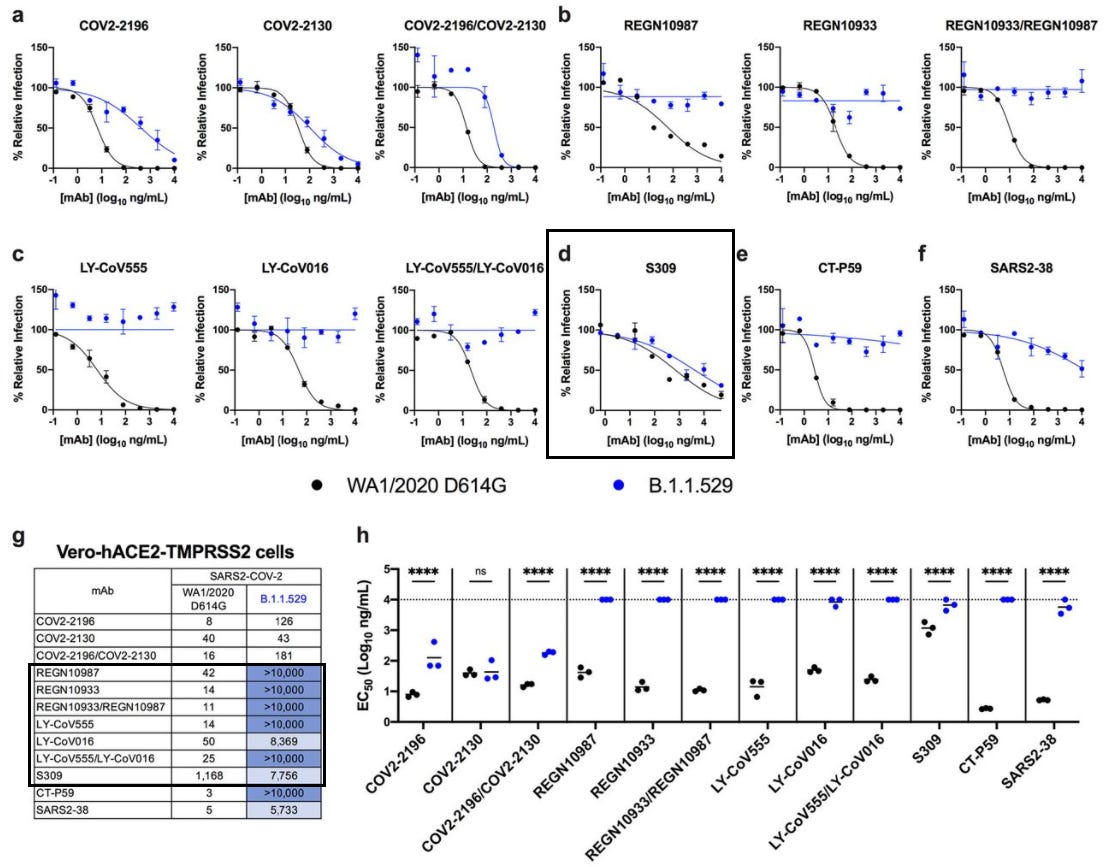
Although not included, it’s important to note a huge distinction between the Vero-TMPRSS2 assay results and the Vero-hACE2-TMPRSS2 results for Sotrovimab, such that the former saw an EC50 (concentration where 50% of neutralization occurs) value around 373 ng/mL while the latter saw an EC50 value of 7,756 ng/mL. Such a stark difference brings into question the important of the cell assay being used and what methodologies are being utilized. In this case, the expression of hACEII by cells play a critical factor in viral binding, altering the effective concentration needed to inhibit antigen/receptor binding.
So far we’ve found plenty of nonclinical evidence, all of which point to the notion that both Eli Lilly and Regeneron’s dual therapies are likely to be ineffective against Omicron, while Sotrovimab is likely to be the best, most effective candidate.
However, the discrepancy in results between cells lines that either did or did not express hACEII receptors indicates another variable that is important to examine in these studies.
Similar results can be found in a preprint from Tada et. al., which also used a cell line that expressed hACEII receptors, and found a marked reduction of Sotrovimab effectiveness in the face of Omicron.
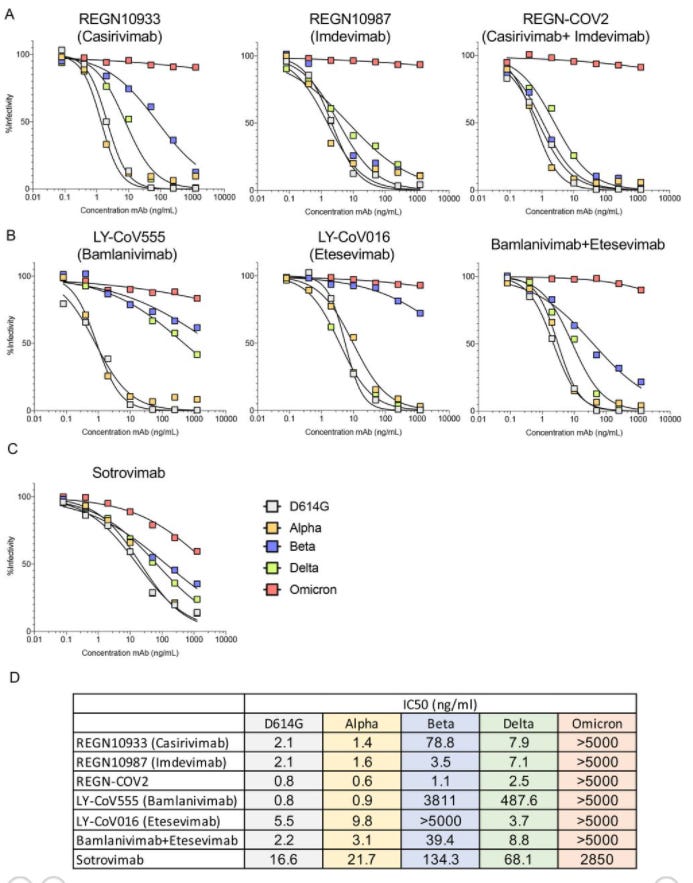
The noticeable difference in Sotrovimab compared to the other studies may point to the cell lines that were used. ACEII receptors are vital to allow SARS-COV2 to bind to host cell attachment and viral entry. Lack of expression in tested cell lines may alter the molecular dynamics and may not actively translate into the real molecular dynamics seen in a natural infection.
Heterogeniety in nonclinical assays need to be taken into considerations, as they may bias the results seen here. More interestingly, the cell line used in the Wilhelm et. al. study also expressed ACEII, and although the results suggested strongly retained effectiveness of Sotrovimab the cell line used was different, adding another possible variable into the mix.
It’s also worth considering that not all Omicron SARS-COV2 isolates may encode for the same mutations. As such, differences in assay results may also be attributed to differences in variant mutations.
Remarks made in the Cao et. al. preprint suggests possible mutations that could reduce Sotrovimab’s effectiveness, which may be what we are seeing here:
VIR-7831 retains strong RBD binding capability, although G339 is part of its epitope, the G339D mutation in Omicron does not appear to affect VIR-7831’s binding; however, VIR-7831’s IC50 is reduced to 181 ng/mL, and may be subject to further reduction against Omicron with R346K.
However, this R346K mutation doesn’t seem to be found in the Omicron variant used in the Tada et. al. study. This brings into question if the use of Omicron variants with different mutations may alter the results of these binding assays. Most researchers are transparent about the mutations found in their form of the Omicron variant that they use in their studies, but possible mutations when incubating viruses may likely cause heterogeneous results to arise, leading to variations in antibody/antigen binding.
Luckily, the Tada et. al. researchers conducted another assay utilizing individual mutations and examining their effects on monoclonal antibody binding.

These results help indicate pivotal amino acids responsible for antibody binding to the antigen. As noted, a few amino acids (such as Imdevimab when confronted with a G448S mutation) led to extremely great loss of antibody binding, meaning that far more antibodies would be needed to elicit an inhibitory effect as noted by the extremely high IC50 value.
These results also indicate the importance and precarious nature of the antigen/antibody binding relationship. This can be seen by the models on the bottom, relating the alteration in binding between the wildtype and Omicron. Although there’s a lot to discuss, I’ll highlight the effects some of these mutations had on Regeneron, as was raised by the researchers. When examining this excerpt, know that the notation of a letter followed by two numbers refers to the epitope of the antibody in question (i.e. the Y53 notation indicates that the amino acid is a tyrosine residue for the heavy chain of Casirivimab at position 53):
The efficacy of Casirivimab (REGN10933) is compromised by mutations K417N, E484A, and Q493K. In the structure of the Casirivimab Fab-spike protein, these mutations are situated in the interface with the Fab heavy chain (Figure 3C). K417N would cause a loss of hydrogen bonding with T28 and T102 of the heavy chain, as well as the loss of a favorable electrostatic interaction with D31 of the heavy chain. E484A would result in the loss of hydrogen bonding with Y53 and S56 of the heavy chain, and Q493K would result in the loss of hydrogen bonding with N74 of the heavy chain.
For Imdevimab (REGN10987), four mutations in the Omicron spike protein lead to significant loss of efficacy: S371L, S373P, N440K, and G446S. The N440K mutation would create steric clashes between K440 and the heavy and light chains and result in charge repulsion with K55 of the light chain. Mutation of G446 to any other (larger) residue (e.g., G446S) would cause a steric clash with N57 of the heavy chain. Mutation of amino acids S371 and S373 adversely affect antibody activity but do not directly contact the Fab (Figure 3C); mutation of these amino acids could alter the stability of this loop segment, affecting the conformation of the nearby region (N440) of antibody binding.
The totality, as outlined above, is what makes or breaks an antibody’s neutralizing capabilities. But the results above run counter to the evidence provided by Chen et. al.’s preprint. In fact, some of the mutations outlined in that paper don’t align with the same deleterious effects from the Tada et. al. preprint. As I stated, the BFE and modeling done by the Chen et. al. study depend on a variety of variables, so biases may be instilled into the modeling software. The Tada et. al. study also compared changes on an individual mutation basis, and so the actual binding of antibodies would be far more different with multiple mutations affecting the structure and conformation of Omicron’s spike.
There’s a lot to speculate about the discrepancies found between those two studies, but it highlights the importance of understanding the heterogeneity between studies. As we can see, the difference in cell lines used, expression of hACEII, incubation and assay methodology all contribute their own biases.
Nonetheless, the evidence is mounting that both Eli Lilly and Regeneron’s dual therapies are the most likely to not prove effective against Omicron. With Sotrovimab, the loss of effectiveness varies by studies, although the overall results suggest that Sotrovimab should hold up to the mutations of Omicron and still confer neutralizing activities against Omicron’s spike. All of these studies were conducted in a nonclinical setting, and as many of the researchers noted the translation into a clinical setting is up for debate. Until clinical studies are conducted we will not know the real-world effects these mutations have on monoclonal therapeutics.




> if monoclonal antibodies such as Regeneron prove ineffective against Omicron, is it still important to provide monoclonals as early treatment in the off chance that the infection could be Delta?
That would be counter-productive if your objective extend the "emergency". ;)
I've been reading that some states have been running low on monoclonal antibody treatment. But maybe it won't matter so much, if they're not as effective.